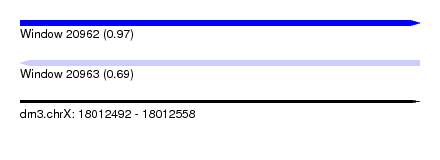
| Sequence ID | dm3.chrX |
|---|---|
| Location | 18,012,492 – 18,012,558 |
| Length | 66 |
| Max. P | 0.972180 |

| Location | 18,012,492 – 18,012,558 |
|---|---|
| Length | 66 |
| Sequences | 4 |
| Columns | 70 |
| Reading direction | forward |
| Mean pairwise identity | 88.94 |
| Shannon entropy | 0.17034 |
| G+C content | 0.31978 |
| Mean single sequence MFE | -17.51 |
| Consensus MFE | -12.60 |
| Energy contribution | -12.72 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.21 |
| Mean z-score | -3.04 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.86 |
| SVM RNA-class probability | 0.972180 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
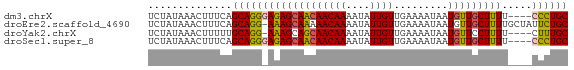
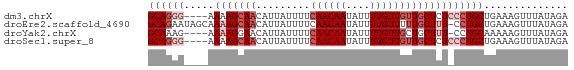
>dm3.chrX 18012492 66 + 22422827 UCUAUAAACUUUCAGCAGGGAGAGCAACAACAAAAUAUUGUUGAAAAUAAUGUUGCUUUU----CCCUGC ..............(((((((((((((((((((....)))).........)))))).)))----)))))) ( -21.80, z-score = -4.79, R) >droEre2.scaffold_4690 8340459 69 + 18748788 UCUAUAAACUUUCAGCAGG-AAAGCAAAAACAAAAUAUUGUUGAAAAUAAUGUUGCUUUUGCUAUUCUGC ..............(((((-(.(((((((....((((((((.....))))))))..))))))).)))))) ( -18.30, z-score = -2.94, R) >droYak2.chrX 16636611 65 + 21770863 UCUAUAAACUUUUUGCAGG-AAAGCAGCAACAAAAUAUUGUUGAAAAUAAUGUUCCUUUU----CUUUGC ................(((-((.....((((((....)))))).........)))))...----...... ( -8.14, z-score = 0.38, R) >droSec1.super_8 317731 66 + 3762037 UCUAUAAACUUUCAGCAGGGAGAGCAACAACAAAAUAUUGUUGAAAAUAAUGUUGCUUUU----CCCUGC ..............(((((((((((((((((((....)))).........)))))).)))----)))))) ( -21.80, z-score = -4.79, R) >consensus UCUAUAAACUUUCAGCAGG_AAAGCAACAACAAAAUAUUGUUGAAAAUAAUGUUGCUUUU____CCCUGC ..............(((((((((((((((((((....)))).........))))))))).....)))))) (-12.60 = -12.72 + 0.12)
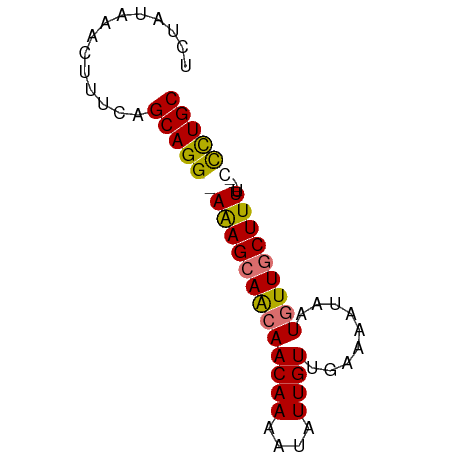
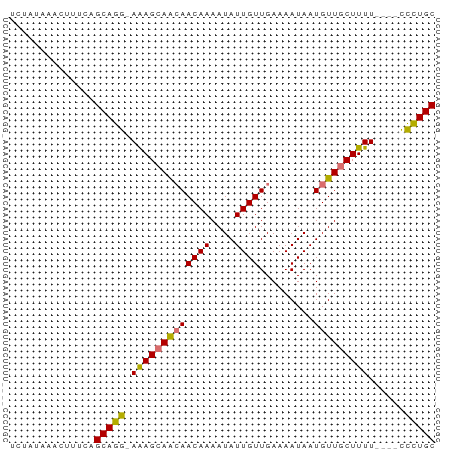
| Location | 18,012,492 – 18,012,558 |
|---|---|
| Length | 66 |
| Sequences | 4 |
| Columns | 70 |
| Reading direction | reverse |
| Mean pairwise identity | 88.94 |
| Shannon entropy | 0.17034 |
| G+C content | 0.31978 |
| Mean single sequence MFE | -17.65 |
| Consensus MFE | -10.34 |
| Energy contribution | -11.90 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.72 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.687641 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
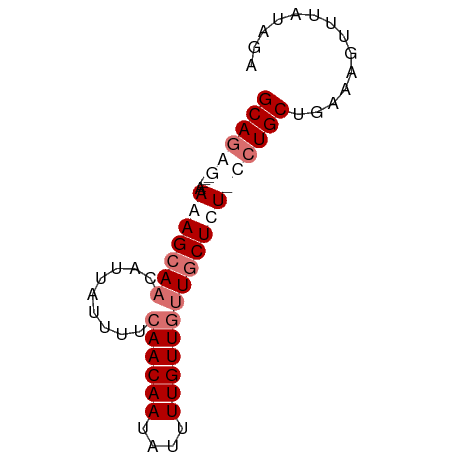
>dm3.chrX 18012492 66 - 22422827 GCAGGG----AAAAGCAACAUUAUUUUCAACAAUAUUUUGUUGUUGCUCUCCCUGCUGAAAGUUUAUAGA ((((((----(..(((((((.........((((....))))))))))).))))))).............. ( -21.70, z-score = -4.25, R) >droEre2.scaffold_4690 8340459 69 - 18748788 GCAGAAUAGCAAAAGCAACAUUAUUUUCAACAAUAUUUUGUUUUUGCUUU-CCUGCUGAAAGUUUAUAGA ((((...(((((((((((...((((......))))..)))))))))))..-.)))).............. ( -18.20, z-score = -2.68, R) >droYak2.chrX 16636611 65 - 21770863 GCAAAG----AAAAGGAACAUUAUUUUCAACAAUAUUUUGUUGCUGCUUU-CCUGCAAAAAGUUUAUAGA ......----.....((((........((((((....)))))).(((...-...)))....))))..... ( -9.00, z-score = 0.29, R) >droSec1.super_8 317731 66 - 3762037 GCAGGG----AAAAGCAACAUUAUUUUCAACAAUAUUUUGUUGUUGCUCUCCCUGCUGAAAGUUUAUAGA ((((((----(..(((((((.........((((....))))))))))).))))))).............. ( -21.70, z-score = -4.25, R) >consensus GCAGAG____AAAAGCAACAUUAUUUUCAACAAUAUUUUGUUGUUGCUCU_CCUGCUGAAAGUUUAUAGA ((((((.......(((((((.((((......))))...))))))).....)))))).............. (-10.34 = -11.90 + 1.56)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:55:09 2011