
| Sequence ID | dm3.chrX |
|---|---|
| Location | 18,007,461 – 18,007,611 |
| Length | 150 |
| Max. P | 0.793750 |

| Location | 18,007,461 – 18,007,611 |
|---|---|
| Length | 150 |
| Sequences | 10 |
| Columns | 172 |
| Reading direction | reverse |
| Mean pairwise identity | 83.57 |
| Shannon entropy | 0.34154 |
| G+C content | 0.53023 |
| Mean single sequence MFE | -48.64 |
| Consensus MFE | -34.62 |
| Energy contribution | -35.51 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.793750 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 18007461 150 - 22422827 ---------CAGGCUAAGAAGGCUAAACAGGCUGAGAUCGAGCGCAAGCGUGCUGAGGUGCGCAAGCGCAUGGAGGAAGCCUCCAAGGCCAAGAAGGCCAAGAAGGGUUUCAUGACCCCAGAGAGGAAGAAGAAACUCAGGGUAAGUUUCCAGCAUAUA------------- ---------..((((.....))))......((((.((....((....))...(((((((((....)))).((((((...)))))).((((.....)))).....(((((....))))).................)))))........)))))).....------------- ( -52.20, z-score = -2.28, R) >droWil1.scaffold_181096 12015069 163 - 12416693 ---------CAGGCUAAGAAGGCUAAACAGGCUGAGAUCGAGCGCAAGCGUGCUGAGGUGCGCAAGCGCAUGGAAGAAGCCUCCAAGGCCAAGAAGGCCAAGAAGGGUUUCAUGACACCAGAAAGGAAGAAGAAACUCAGGGUAAGUUUCCUCCCCCCCCACCCCAUCAAUA ---------..((((.....((((....(((((....(((.((((..(((..(....)..)))..)))).)))....)))))....)))).....))))..((.(((((((.((....))))))((..((.((((((.......)))))).))..)).....))).)).... ( -52.00, z-score = -1.02, R) >droSec1.super_8 312826 150 - 3762037 ---------CAGGCUAAGAAGGCUAAACAGGCUGAGAUCGAGGGCAAGCGGGCUGAGAUGCCCAAGCGCAUGGAGGAAGCCUCCAAGGCCAAGAAGGCCAAGAAGGCUUUCAUGACCCCAGAGAGGAAGAAGAAACUCAGGGUAAGUAUACUGCAUAUA------------- ---------(((.....(((((((.....((((....((...(((..((((((......))))..))...((((((...))))))..)))..)).)))).....)))))))...((((....(((..........))).)))).......)))......------------- ( -47.00, z-score = -0.87, R) >droSim1.chrX_random 4622929 150 - 5698898 ---------CAGGCUAAGAAGGCUAAACAGGCUGAGAUCGAGCGCAAGCGUGCUGAGGUGCGCAAGCGCAUGGAGGAAGCCUCCAAGGCCAAGAAGGCCAAGAAGGGUUUCAUGACCCCAGAGAGGAAGAAGAAACUCAGGGUAAGUUAUCUGCAUAUA------------- ---------...((..(((..(((..((...(((((.((..((((..(((..(....)..)))..)))).((((((...)))))).((((.....))))..)).(((((....))))).................))))).)).)))..))))).....------------- ( -51.10, z-score = -1.96, R) >apiMel3.Group2 1904786 152 - 14465785 CGGAAAAUGUAGGAAAAGAAGAGGAAACAGGCGGAGACGGACAGAAAGAGGGCGGAGGUCCGCGCCCGUCUCGAAGAGGCUUCCAAAGCCAAGAAGGCGAAGAAGGGUUUCAUGACCCCUGACAGGAAGAAGAAGCUUAGGGUAAGUUUCCA-------------------- ......................((((((..((.(((((((.(.....).(.((((....)))).))))))))....(((((((....(((.....)))......(((((....))))).............)))))))...))..)))))).-------------------- ( -48.60, z-score = -1.71, R) >anoGam1.chrU 38422685 148 - 59568033 ----------AAGCGAAGAAGGCUAAGCAGGCGGAGAUCGAGCGUAAGCGUGCUGAAGUAAGGAAGCGUAUGGAAGAGGCUUCUAAGGCAAAGAAGGCUAAGAAGGGUUUCAUGACCCCUGAACGCAAGAAGAAACUGAGGGUAAGUAUUAAAAACAU-------------- ----------..(((.....((((.((((.((...(......)....)).))))...((..((((((.(.......).))))))...))......)))).....(((((....))))).....)))........(((.......)))...........-------------- ( -31.60, z-score = 0.74, R) >droYak2.chrX 16631747 149 - 21770863 ---------CAGGCUAAGAAGGCUAAACAGGCUGAGAUCGAGCGCAAGCGUGCUGAGGUGCGCAAGCGCAUGGAGGAAGCCUCCAAGGCCAAGAAGGCCAAGAAGGGUUUCAUGACUCCAGAGAGGAAGAAGAAACUCAGGGUAAGUUGUACCAGCUC-------------- ---------..((((.....((((.....((((........((((..(((..(....)..)))..)))).((((((...)))))).)))).....)))).....(((((((.(..(((....)))..)...)))))))..((((.....)))))))).-------------- ( -53.60, z-score = -2.22, R) >droEre2.scaffold_4690 8335530 142 - 18748788 ---------CAGGCUAAGAAGGCUAAACAGGCUGAGAUCGAGCGCAAGCGUGCUGAGGUGCGCAAGCGCAUGGAGGAAGCCUCCAAGGCCAAGAAGGCCAAGAAGGGUUUCAUGACCCCAGAGAGGAAGAAGAAACUCAGGGUAAGUUUUC--------------------- ---------........(((((((..((...(((((.((..((((..(((..(....)..)))..)))).((((((...)))))).((((.....))))..)).(((((....))))).................))))).)).)))))))--------------------- ( -50.40, z-score = -2.32, R) >droAna3.scaffold_13335 1714298 150 + 3335858 ---------CAGGCUAAGAAGGCUAAACAGGCUGAGAUCGAGCGCAAGCGUGCUGAGGUGCGCAAGCGCAUGGAGGAAGCCUCCAAGGCCAAGAAGGCCAAGAAGGGUUUCAUGACCCCAGAGAGGAAGAAGAAACUCAGGGUGAGUUACGAUUACAAG------------- ---------..((((.....))))........((.(((((.((....))...(((((((((....)))).((((((...)))))).((((.....)))).....(((((....))))).................))))).........))))).))..------------- ( -50.90, z-score = -2.46, R) >droGri2.scaffold_15203 8432819 138 - 11997470 ---------CAGGCUAAGAAGGCUAAACAGGCUGAGAUCGAGCGCAAGCGUGCUGAGGUGCGCAAGCGCAUGGAGGAAGCCUCUAAGGCCAAGAAGGCCAAGAAGGGUUUCAUGACCCCAGAGAGGAAGAAGAAACUCCGGGUAAGU------------------------- ---------..((((.....))))......(((...(((..((((..(((..(....)..)))..)))).(((((....(((((..((((.....)))).....(((((....)))))...))))).........)))))))).)))------------------------- ( -49.02, z-score = -2.15, R) >consensus _________CAGGCUAAGAAGGCUAAACAGGCUGAGAUCGAGCGCAAGCGUGCUGAGGUGCGCAAGCGCAUGGAGGAAGCCUCCAAGGCCAAGAAGGCCAAGAAGGGUUUCAUGACCCCAGAGAGGAAGAAGAAACUCAGGGUAAGUUUCCA_CACAU______________ ...........((((.....))))....(((((....(((.((((..((((((....))))))..)))).)))....)))))((..((((.....)))).....(((((((......((.....)).....))))))).))............................... (-34.62 = -35.51 + 0.89)

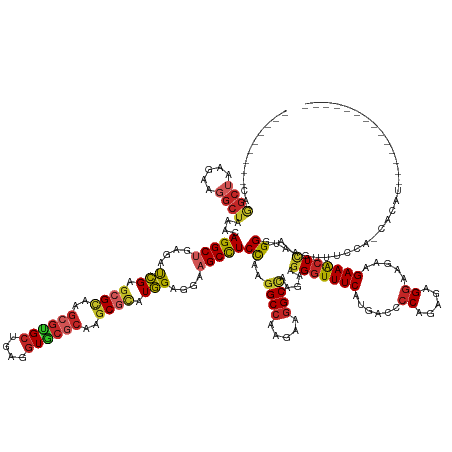
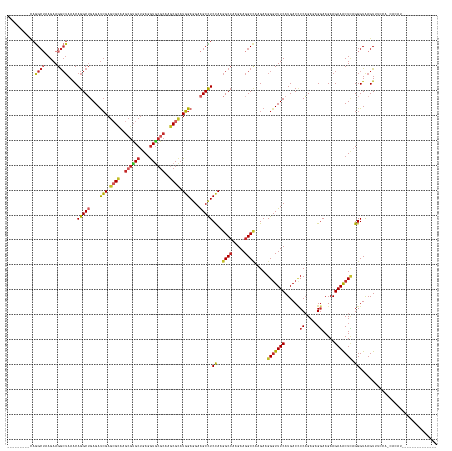
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:55:08 2011