| Sequence ID | dm3.chrX |
|---|---|
| Location | 17,986,955 – 17,987,059 |
| Length | 104 |
| Max. P | 0.997752 |

| Location | 17,986,955 – 17,987,059 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 67.88 |
| Shannon entropy | 0.54510 |
| G+C content | 0.32594 |
| Mean single sequence MFE | -21.52 |
| Consensus MFE | -14.46 |
| Energy contribution | -15.02 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.17 |
| SVM RNA-class probability | 0.997752 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
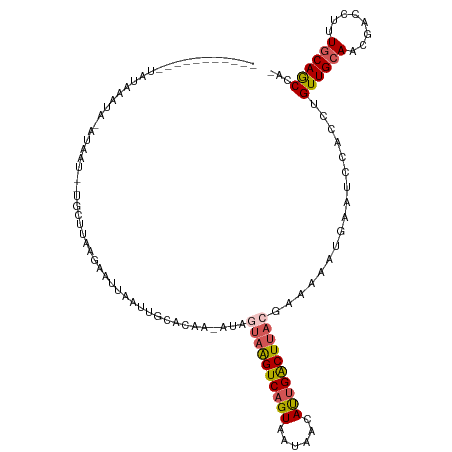
>dm3.chrX 17986955 104 - 22422827 -----------UAUAAAUACAUGGU--UGCUUAAGAAUUAAUUGCACAA-AUAGUAAGUCAGUAAUAAAAUUGACUUACAAAAAAUUAAUCCACCUGUUGCAACGACCUGUGCAGCCA- -----------..........((((--(((...((......(((((...-...((((((((((......))))))))))............(....).)))))....))..)))))))- ( -23.90, z-score = -3.04, R) >droSim1.chrX 13992572 103 - 17042790 -----------UAUAACUA-AGAAU--AAGUUAAGAAUGAAUUGCAUAA-AUAGUAAGUCAGUAAUAACAUUGACUUACGAAAAAUGAAUCCACCUGUUGCAUCGACCUUUGCAGCCA- -----------..(((((.-.....--.)))))....((.(((.(((..-...((((((((((......)))))))))).....)))))).))...((((((........))))))..- ( -21.80, z-score = -3.39, R) >droSec1.super_8 293385 103 - 3762037 -----------UAUAACUA-AGAAU--UGCUUAAGAAUUAAUGGCACAA-AUAGUAAGUCAGUAAUAACAUUGACUUACGAAAAAUAAAUCCACCUGUUGCAUCGACCUUUGCAGCAA- -----------........-.....--((((...........))))...-...((((((((((......))))))))))................(((((((........))))))).- ( -22.70, z-score = -3.24, R) >droYak2.chrX 16611995 91 - 21770863 -----------UACAAAUACAUGUU--UGCUUUAGAAUUAAUUCUACAA-AUAUUAAGUCUGUAAUAAUAUUGACUU--------------CACCUGUUGCAAUGGACACUGCAGCCCA -----------.........(((((--((...(((((....))))))))-)))).(((((.(((....))).)))))--------------.....((((((........))))))... ( -15.10, z-score = -0.85, R) >droEre2.scaffold_4690 8315670 119 - 18748788 UGCAAUGUAUUUGGUAUUUGAUAUUGAUACUUUAGAAGCAAUUUGACAACAAUUUUGGUCAGUAGUAAUACUGGCUUAUCGAACAUGAACCAACCUGUUGCAAUCAGCUUUCCAACCCA (((((((...(((((...((...((((((.....(((....)))............((((((((....)))))))))))))).))...)))))..)))))))................. ( -24.10, z-score = -0.62, R) >consensus ___________UAUAAAUA_AUAAU__UGCUUAAGAAUUAAUUGCACAA_AUAGUAAGUCAGUAAUAACAUUGACUUACGAAAAAUGAAUCCACCUGUUGCAACGACCUUUGCAGCCA_ .....................................................((((((((((......)))))))))).................((((((........))))))... (-14.46 = -15.02 + 0.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:55:07 2011