
| Sequence ID | dm3.chrX |
|---|---|
| Location | 17,933,183 – 17,933,303 |
| Length | 120 |
| Max. P | 0.732380 |

| Location | 17,933,183 – 17,933,303 |
|---|---|
| Length | 120 |
| Sequences | 11 |
| Columns | 124 |
| Reading direction | reverse |
| Mean pairwise identity | 78.44 |
| Shannon entropy | 0.45670 |
| G+C content | 0.55068 |
| Mean single sequence MFE | -39.82 |
| Consensus MFE | -27.93 |
| Energy contribution | -28.75 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.02 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.732380 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 17933183 120 - 22422827 UUUCUUCGAUCGGAACCGGAUUUGGG----AAACAGCCGCCAAGAUGACCAUGUGGCAGAGUGGCGGCAUGGGAGGCCAUGGCUCCCAGAACAAUCCAUGGAUGAAGCUGAUGGGCAUCGUCCA .((((......))))..(((((.((.----...).(((((((...((.((....))))...))))))).((((((.(....)))))))...)))))).(((((((.(((....))).))))))) ( -49.00, z-score = -2.11, R) >droSec1.super_8 239787 120 - 3762037 UUUCUUCGAUCGGAACCGGAUUUGGG----AAACAGCCGCCAAGAUGACCAUGUGGCAGAGUGGCGGCAUGGGAGGCCAUGGCUCCCAGAACAAUCCAUGGAUGAAGCUGAUGGGCAUCGUACA .(((.((.((.(((....(.((((((----(..(.(((((((...((.((....))))...)))))))((((....)))))..))))))).)..))))).)).)))(((....)))........ ( -42.30, z-score = -0.82, R) >droYak2.chrX 16553517 120 - 21770863 UUUCUUCGAUCGGAAACGGAUUUGGG----AAACAGCCGCCAAGAUGACCAUGUGGCAGAGUGGCGGCAUGGGAGGCCAUGGCUCCCAGAACAAUCCAUGGAUGAAGCUGAUGGGCAUCGUCCA (((((..(((((....).))))..))----)))..(((((((...((.((....))))...))))))).((((((.(....)))))))..........(((((((.(((....))).))))))) ( -50.90, z-score = -2.94, R) >droEre2.scaffold_4690 8261590 120 - 18748788 UUUCUUCGAUCGGAACCGGAUUCGGG----AAACAGCCGCCAAGAUGACCAUGUGGCAGAGUGGCGGCAUGGGAGGCCAUGGCUCCCAGAACAAUCCAUGGAUGAAGCUGAUGGGCAUCGUUCA ......((((((....)))..)))((----(....(((((((...((.((....))))...))))))).((((((.(....)))))))......))).(((((((.(((....))).))))))) ( -46.50, z-score = -1.56, R) >droAna3.scaffold_13335 1642652 92 + 3335858 --------------------------------ACAGCCGCCAAGAUGACCAUGUGGCAGAGUGGCGGCAUGGGAGGCCAUGGCUCCCAGAACAAUCCAUGGAUGAAGCUGAUGGGCAUUGUCCA --------------------------------...(((((((...((.((....))))...))))))).((((((.(....)))))))..........((((..(.(((....))).)..)))) ( -39.70, z-score = -2.08, R) >droPer1.super_21 190011 105 - 1645244 --------------UCAGAGUGC-AA----AGAGAGCCGCCAAGAUGACUAUGUGGCAGAGUGGCGGCAUGGGAGGCCAUGGCUCCCAGAACAAUCCAUGGAUGAAGCUGAUGGGCAUCGUGCA --------------.....(..(-..----.....(((((((...((.((....))))...))))))).((((((.(....))))))).....(((....)))((.(((....))).)))..). ( -35.90, z-score = -0.51, R) >dp4.chrXL_group1e 5279079 105 - 12523060 --------------UCAGAGUGC-AA----AGAGAGCCGCCAAGAUGACUAUGUGGCAGAGUGGCGGCAUGGGAGGCCAUGGCUCCCAGAACAAUCCAUGGAUGAAGCUGAUGGGCAUCGUGCA --------------.....(..(-..----.....(((((((...((.((....))))...))))))).((((((.(....))))))).....(((....)))((.(((....))).)))..). ( -35.90, z-score = -0.51, R) >droWil1.scaffold_181150 1920183 115 + 4952429 -----UUAAAAGGAUUAAAGGGUAAA----AAAAAGCCGCCAAGAUGACCAUGUGGCAGAGUGGUGGUAUGGGAGGCCAUGGCUCCCAAAAUAAUCCAUGGAUGAAACUAAUGGGCAUUGUCCA -----......((((((.........----.....(((((((...((.((....))))...))))))).((((((.(....)))))))...)))))).((((..(..((....))..)..)))) ( -38.50, z-score = -2.60, R) >droMoj3.scaffold_6473 2421558 93 + 16943266 -------------------------------ACCAAACGCCAAGAUGACGAUGUGGCAGAGUGGCGGCAUGGGAGGCCAUGGCUCCCAGAACAAUCCAUGGAUGAAGCUGAUGGGCAUCGUGCA -------------------------------......(((((...((.(......)))...)))))((((((..(.((((((((.(((..........)))....)))).)))).).)))))). ( -28.80, z-score = 0.50, R) >droSim1.chrX 13969232 120 - 17042790 UUUCUUCGAUCGGAACCGGAUUUGGG----AAACAGCCGCCAAGAUGACCAUGUGGCAGAGUGGCGGCAUGGGAGGCCAUGGCUCCCAGAACAAUCCAUGGAUGAAGCUGAUGGGCAUCGUACA .(((.((.((.(((....(.((((((----(..(.(((((((...((.((....))))...)))))))((((....)))))..))))))).)..))))).)).)))(((....)))........ ( -42.30, z-score = -0.82, R) >triCas2.ChLG3 22848330 116 + 32080666 -----UUUUUUCGGUCGGGGGGUAAAUGGUGAUGAUCCGCAAUCAUGACGAUGUGGCAGAGUGG---GAUGGGGGGGCCCGGGGGCAAGAACAACGCGUGGAUGAAGCUGAUGGGGAUCGUCCA -----...(..((.((....(.((.((.((.(((((.....))))).)).)).)).).)).)).---.)((((.((.((((..(((..(..((.....))..)...)))..))))..)).)))) ( -28.20, z-score = 2.21, R) >consensus _____UCGAUCGGAACCGGAUUUGGG____AAACAGCCGCCAAGAUGACCAUGUGGCAGAGUGGCGGCAUGGGAGGCCAUGGCUCCCAGAACAAUCCAUGGAUGAAGCUGAUGGGCAUCGUCCA ...................................(((((((...((.((....))))...))))))).((((((.(....)))))))..........((.((((.(((....))).)))).)) (-27.93 = -28.75 + 0.81)
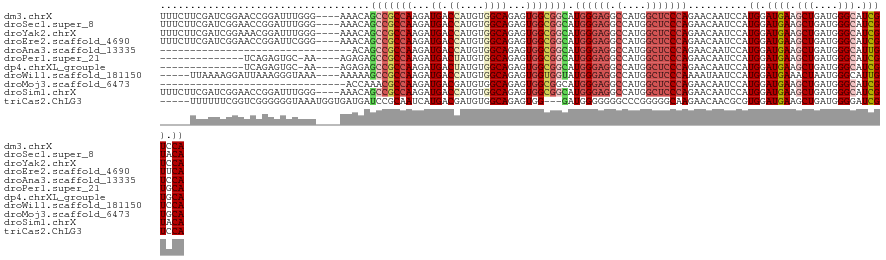
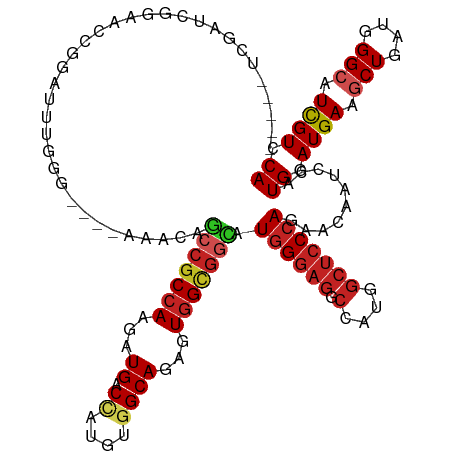

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:55:01 2011