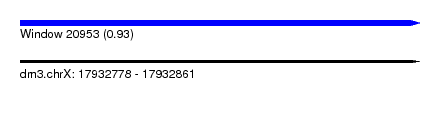
| Sequence ID | dm3.chrX |
|---|---|
| Location | 17,932,778 – 17,932,861 |
| Length | 83 |
| Max. P | 0.930867 |

| Location | 17,932,778 – 17,932,861 |
|---|---|
| Length | 83 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 48.39 |
| Shannon entropy | 0.85342 |
| G+C content | 0.40668 |
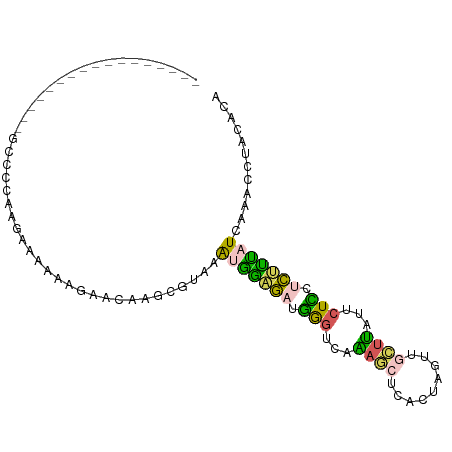
| Mean single sequence MFE | -14.90 |
| Consensus MFE | -6.60 |
| Energy contribution | -5.88 |
| Covariance contribution | -0.72 |
| Combinations/Pair | 1.93 |
| Mean z-score | -0.85 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.930867 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
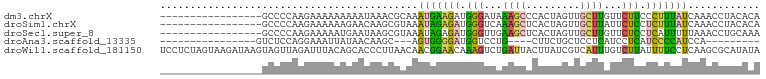
>dm3.chrX 17932778 83 + 22422827 -----------------GCCCCAAGAAAAAAAAAUAAACGCAAAUGAAGAUGGGAUAAAGCCCACUAGUUGCUUGUUCUUCCCUUUAUCAAACCUACACA -----------------...........................(((.((.((((..((((.........)))).....)))).)).))).......... ( -10.10, z-score = -0.20, R) >droSim1.chrX 13960197 83 + 17042790 -----------------GCCCCAAGAAAAAAGAACAAGCGUAAAUAGAGAUGGGUCAAAGCUCACUAGUUGCUUAUUCUCCUCUUUAUCAAACCUACACA -----------------.....((((....((((.(((((....(((...(((((....))))))))..))))).))))..))))............... ( -12.30, z-score = -0.33, R) >droSec1.super_8 239152 83 + 3762037 -----------------GCCCCAAGAAAAAUGAAUAAGCGUAAAUAGAGAUGGGUUGAAGCUCACUAGUUGCUUGUUCUCCUCAUUUUUAAACCUGCAAA -----------------((......(((((((((((((((....(((...(((((....))))))))..))))))).....))))))))......))... ( -17.50, z-score = -1.31, R) >droAna3.scaffold_13335 1641993 68 - 3335858 ----------------GUCUCCAGGAAAUUAUAACAAGC---AGUGGGGAUGGUCCUG----CUUCUGCUCCUCAUCCUCAUCCCCAUCCA--------- ----------------.......(((.......((....---.))((((((((....(----(....)).........)))))))).))).--------- ( -15.42, z-score = -0.18, R) >droWil1.scaffold_181150 3035606 100 + 4952429 UCCUCUAGUAAGAUAAGUAGUUAGAUUUACAGCACCCUUAACAACGGAACAAAGUCUGAUUACUUAUCGUCAUUUGUCUUAUUUUCCUCAAGCGCAUAUA ...........(((((((((((((((((.......((........))....)))))))))))))))))................................ ( -19.20, z-score = -2.21, R) >consensus _________________GCCCCAAGAAAAAAGAACAAGCGUAAAUGGAGAUGGGUCAAAGCUCACUAGUUGCUUAUUCUCCUCUUUAUCAAACCUACACA ...........................................(((((((.(((...((((.........))))...))).)))))))............ ( -6.60 = -5.88 + -0.72)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:55:00 2011