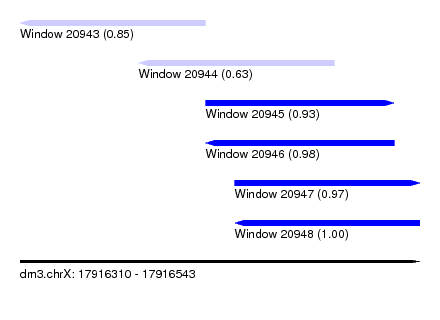
| Sequence ID | dm3.chrX |
|---|---|
| Location | 17,916,310 – 17,916,543 |
| Length | 233 |
| Max. P | 0.997301 |

| Location | 17,916,310 – 17,916,418 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 79.14 |
| Shannon entropy | 0.40291 |
| G+C content | 0.50095 |
| Mean single sequence MFE | -37.77 |
| Consensus MFE | -22.11 |
| Energy contribution | -26.12 |
| Covariance contribution | 4.00 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.846060 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
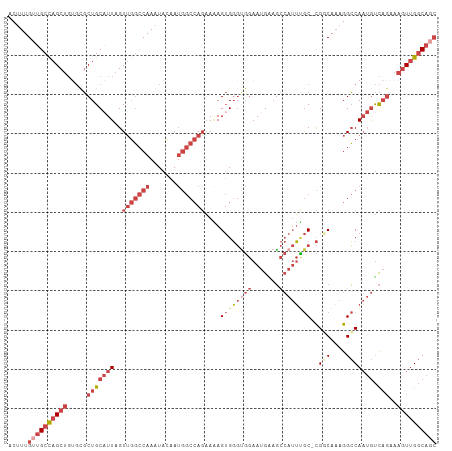
>dm3.chrX 17916310 108 - 22422827 ACUUUGUUGCCAGCUGUGCGCUGCAUUUGUUGGCCAAGUACAAUGGCCAGAAAAGUGGGUGGAAUGAAGCCAUUCGC-CGGCAAAGGCCAAUGUCAGAAAGUUGGCAGC .....((((((((((.....(((((((..(((((((.......)))))))....((((((((.......))))))))-.(((....))))))).)))..)))))))))) ( -43.10, z-score = -2.29, R) >droAna3.scaffold_13335 1623826 101 + 3335858 ACUUUG--GCUAGCU----CCUGCAUUAGUAACACAAAUACUAACACAAUCACAGAGCCACAAAAGGCUAAAAGGACAUGUCAAAGGGCACUACUC--CUCUCGACAGC .(((((--((....(----(((...((((((.......))))))...........((((......))))...))))...)))))))..........--........... ( -20.80, z-score = -1.33, R) >droEre2.scaffold_4690 8245682 108 - 18748788 ACCUUGUUGCCAGCUGUGCGCUGCAUUAGUUGGCCAAGUACAAUGGCCAGAAGAGUGGGCGGAAUGGAGCCAUUUGC-CGUCAAAGGCCAAUGUCAGAAAGUUGGCAGC .....((((((((((...((((.((((..(((((((.......)))))))...)))))))).......(.((((.((-(......))).)))).)....)))))))))) ( -42.00, z-score = -1.90, R) >droYak2.chrX 16536630 107 - 21770863 ACUUUGUUGCCAGCUGCGCGCUGCCUUAGUUGGCCAAGUAUAAUGGCCAGAAAAGUGGGUGGAAUGGAGCCAUUUGC-CGUCACAGGCCAA-GUCAGAAAGUUGGCAGC .....((((((((((....(((((((...(((((((.......)))))))....(((((..((.((....))))..)-)..)))))))..)-)).....)))))))))) ( -39.70, z-score = -1.21, R) >droSec1.super_8 216756 106 - 3762037 ACUUUGUUGCCAGCUGUGUGCUGCAUUAGUUGGCCAAAUACAAUGGCCAGAAAAGUGGGUGGAAUGAAGCCAUUUGC-CGGCAAAGGCCAAUGUCAGAAAGUUGGCA-- .......((((((((.....(((((((..(((((((.......)))))))....(..(((((.......)))))..)-.(((....))))))).)))..))))))))-- ( -38.80, z-score = -1.72, R) >droSim1.chrX 13943926 108 - 17042790 ACUUUGUUGCCAGCUGUGUGCUGCAUUAGUUGGCCAAAUACAAUGGCCAGAAAAGUGGGUGGAAUGAAGCCAUUUGC-CGGCAAAGGCCAAUGUCAGAAAGUUGGCAGC .....((((((((((.....(((((((..(((((((.......)))))))....(..(((((.......)))))..)-.(((....))))))).)))..)))))))))) ( -42.20, z-score = -2.58, R) >consensus ACUUUGUUGCCAGCUGUGCGCUGCAUUAGUUGGCCAAAUACAAUGGCCAGAAAAGUGGGUGGAAUGAAGCCAUUUGC_CGGCAAAGGCCAAUGUCAGAAAGUUGGCAGC .....((((((((((.....(((((((..(((((((.......)))))))....((((((((.......))))))))..(((....))))))).)))..)))))))))) (-22.11 = -26.12 + 4.00)

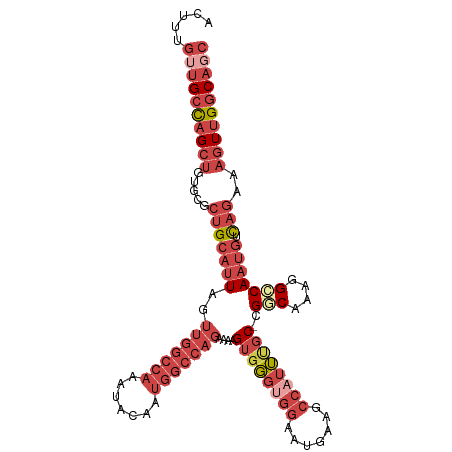
| Location | 17,916,379 – 17,916,493 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 83.75 |
| Shannon entropy | 0.24987 |
| G+C content | 0.46327 |
| Mean single sequence MFE | -28.52 |
| Consensus MFE | -24.94 |
| Energy contribution | -24.25 |
| Covariance contribution | -0.69 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.15 |
| Structure conservation index | 0.87 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.625004 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 17916379 114 - 22422827 CAAAUCGAAACCGAAACCGAAAUCGAAAUCCAAGCUGGACCUGGUCUUGUUCCUAAAUCUGGA-UUUUGACCAUUCACUUUGUUGCCAGCUGUGCGCUGCAUUUGUUGGCCAAGU ....((((...((....))...))))....((((.((((..(((((....(((.......)))-....))))))))).))))..((((((.((((...))))..))))))..... ( -29.40, z-score = -1.38, R) >droEre2.scaffold_4690 8245751 114 - 18748788 CAAAUCAAAAUCGAAAUCGCAAUUGAAAUCCGUGCUGGACCUGGUCUAAUUCCCGAAUCCGGA-UUUUGACCAUUCACCUUGUUGCCAGCUGUGCGCUGCAUUAGUUGGCCAAGU ....((((((((......(((...(....)..)))((((..(((........)))..))))))-))))))........((((..(((((((((((...))).)))))))))))). ( -30.50, z-score = -1.52, R) >droSec1.super_8 216823 103 - 3762037 ------------CAAAUCGAAAUCGAAAUCCGAGCUGGACCUGGUCCUGUUCCUGAAUCUGGGUUUUUGACCAUUCACUUUGUUGCCAGCUGUGUGCUGCAUUAGUUGGCCAAAU ------------....(((....)))....((((.((((..(((((..(..(((......)))..)..))))))))).))))..((((((((((.....)).))))))))..... ( -27.10, z-score = -0.85, R) >droSim1.chrX 13943995 103 - 17042790 ------------CAAAUCGAAAUCGAAAUCCGAGCUGGACCUGGUCCUGUUCCUGAAUCUGGGUUUUUGACCAUUCACUUUGUUGCCAGCUGUGUGCUGCAUUAGUUGGCCAAAU ------------....(((....)))....((((.((((..(((((..(..(((......)))..)..))))))))).))))..((((((((((.....)).))))))))..... ( -27.10, z-score = -0.85, R) >consensus ____________CAAAUCGAAAUCGAAAUCCGAGCUGGACCUGGUCCUGUUCCUGAAUCUGGA_UUUUGACCAUUCACUUUGUUGCCAGCUGUGCGCUGCAUUAGUUGGCCAAAU ........................(((.(((.....)))..(((((....(((.......))).....))))))))..((((..(((((((((((...))).)))))))))))). (-24.94 = -24.25 + -0.69)

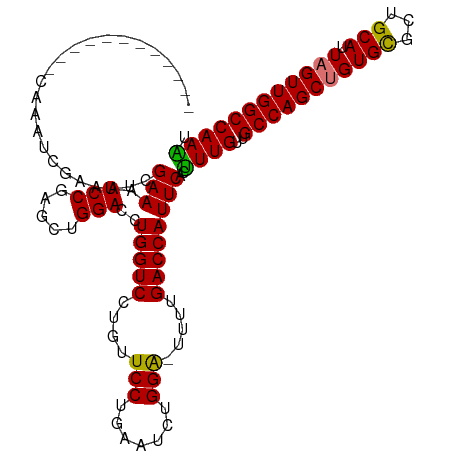
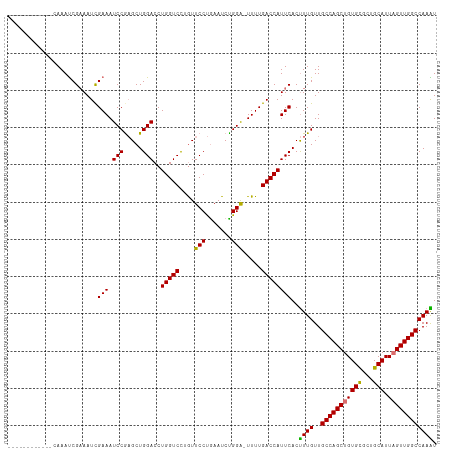
| Location | 17,916,418 – 17,916,528 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 82.49 |
| Shannon entropy | 0.26813 |
| G+C content | 0.49027 |
| Mean single sequence MFE | -35.24 |
| Consensus MFE | -30.27 |
| Energy contribution | -30.46 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.86 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.35 |
| SVM RNA-class probability | 0.929691 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 17916418 110 + 22422827 GAAUGGUCAAAA-UCCAGAUUUAGGAACAAGACCAGGUCCAGCUUGGAUUUCGAUUUCGGUUUCGGUUUCGAUUUGGAGUUGCUGGACCCGGAAUACACGGA-----AUCCGGACU ...(((((....-(((.......)))....)))))(((((.....(((((((((((((((.((((((..(........)..)))))).)))))))...))))-----))))))))) ( -38.40, z-score = -2.24, R) >droEre2.scaffold_4690 8245790 115 + 18748788 GAAUGGUCAAAA-UCCGGAUUCGGGAAUUAGACCAGGUCCAGCACGGAUUUCAAUUGCGAUUUCGAUUUUGAUUUGGAGUCGCUGGACCCGGAACACACGGAUUCUGAUUCGGACU ....((((..((-((.(((((((.........((.((((((((...((((((((...(((........)))..)))))))))))))))).))......))))))).))))..)))) ( -41.26, z-score = -2.77, R) >droSec1.super_8 216862 99 + 3762037 GAAUGGUCAAAAACCCAGAUUCAGGAACAGGACCAGGUCCAGCUCGGAUUUCGAUUUCG------------AUUUGGAGUUGCUGGACCCGGAAUACACGGA-----AUCCGGACU ...(((((.....((........)).....)))))((((((((...((((((((.....------------..))))))))))))))))((((.........-----.)))).... ( -31.50, z-score = -1.16, R) >droSim1.chrX 13944034 99 + 17042790 GAAUGGUCAAAAACCCAGAUUCAGGAACAGGACCAGGUCCAGCUCGGAUUUCGAUUUCG------------AUUUGGAGUUGCUGGACCCGGAAUACACGGA-----AACCGGACU ...(((((.....((........)).....)))))((((((((...((((((((.....------------..)))))))))))))))).........(((.-----..))).... ( -29.80, z-score = -0.89, R) >consensus GAAUGGUCAAAA_CCCAGAUUCAGGAACAAGACCAGGUCCAGCUCGGAUUUCGAUUUCG____________AUUUGGAGUUGCUGGACCCGGAAUACACGGA_____AUCCGGACU ...(((((.....(((.......)))....)))))((((((((...((((((((...................)))))))))))))))).........((((......)))).... (-30.27 = -30.46 + 0.19)

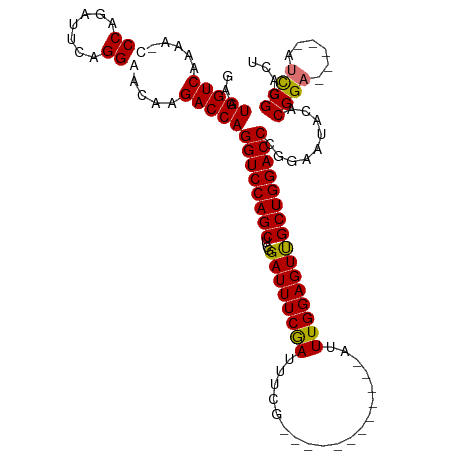
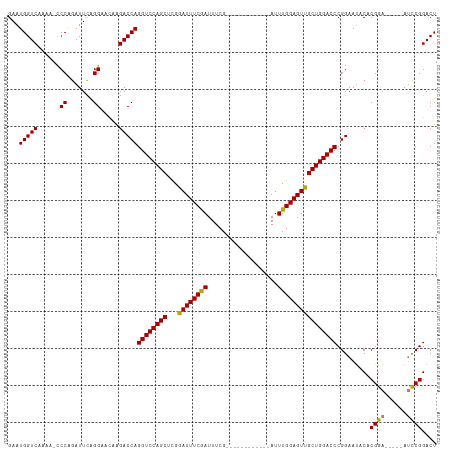
| Location | 17,916,418 – 17,916,528 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 82.49 |
| Shannon entropy | 0.26813 |
| G+C content | 0.49027 |
| Mean single sequence MFE | -34.38 |
| Consensus MFE | -24.95 |
| Energy contribution | -25.02 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.00 |
| SVM RNA-class probability | 0.978660 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 17916418 110 - 22422827 AGUCCGGAU-----UCCGUGUAUUCCGGGUCCAGCAACUCCAAAUCGAAACCGAAACCGAAAUCGAAAUCCAAGCUGGACCUGGUCUUGUUCCUAAAUCUGGA-UUUUGACCAUUC (((((((((-----(..(..((..(((((((((((.........(((....)))...((....))........)))))))))))...))..)...))))))))-)).......... ( -35.60, z-score = -3.26, R) >droEre2.scaffold_4690 8245790 115 - 18748788 AGUCCGAAUCAGAAUCCGUGUGUUCCGGGUCCAGCGACUCCAAAUCAAAAUCGAAAUCGCAAUUGAAAUCCGUGCUGGACCUGGUCUAAUUCCCGAAUCCGGA-UUUUGACCAUUC ........((((((((((..(...((((((((((((........((((...((....))...))))......))))))))))))((........)))..))))-))))))...... ( -37.04, z-score = -3.14, R) >droSec1.super_8 216862 99 - 3762037 AGUCCGGAU-----UCCGUGUAUUCCGGGUCCAGCAACUCCAAAU------------CGAAAUCGAAAUCCGAGCUGGACCUGGUCCUGUUCCUGAAUCUGGGUUUUUGACCAUUC ..(((((((-----((.(.(.((.(((((((((((...((.....------------.))..(((.....))))))))))))))....)).)).)))))))))............. ( -34.70, z-score = -2.73, R) >droSim1.chrX 13944034 99 - 17042790 AGUCCGGUU-----UCCGUGUAUUCCGGGUCCAGCAACUCCAAAU------------CGAAAUCGAAAUCCGAGCUGGACCUGGUCCUGUUCCUGAAUCUGGGUUUUUGACCAUUC ..(((((.(-----((.(.(.((.(((((((((((...((.....------------.))..(((.....))))))))))))))....)).)).))).)))))............. ( -30.20, z-score = -1.49, R) >consensus AGUCCGGAU_____UCCGUGUAUUCCGGGUCCAGCAACUCCAAAU____________CGAAAUCGAAAUCCGAGCUGGACCUGGUCCUGUUCCUGAAUCUGGA_UUUUGACCAUUC ..(((((((...............(((((((((((......................................)))))))))))............)))))))............. (-24.95 = -25.02 + 0.06)

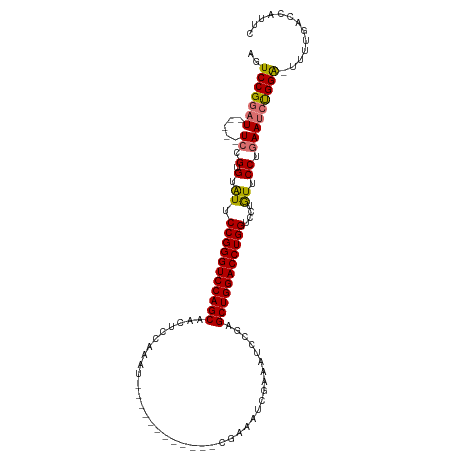
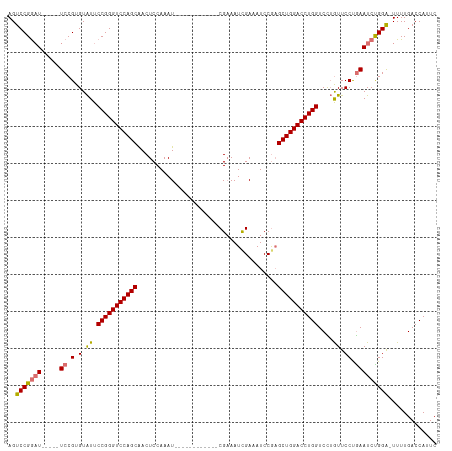
| Location | 17,916,435 – 17,916,543 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 69.78 |
| Shannon entropy | 0.50624 |
| G+C content | 0.50509 |
| Mean single sequence MFE | -33.74 |
| Consensus MFE | -19.58 |
| Energy contribution | -21.83 |
| Covariance contribution | 2.25 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.81 |
| SVM RNA-class probability | 0.968839 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 17916435 108 + 22422827 AUUUAGGAACAAGACCAGGUCCAGCUUGGAUUUCGAUUUCGGUUUCGGUUUCGAUUUGGAGUUGCUGGACCCGGAAUACACGGA-----AUCCGGACUGCAGAUUCCGGAUCC-- ..............((.((((((((...((((((((..((((........)))).)))))))))))))))).)).......(((-----.((((((........)))))))))-- ( -39.20, z-score = -2.26, R) >droAna3.scaffold_13335 1623937 92 - 3335858 AUGCAGGUUCAGGACCUGG--CAUUCC--AUUUCCAUUCCC------------AUUCAGAGU-ACCCAUUCCAAAAUGGAAAAU------CCCGGAUUCCGUAUCCGGAACAUUC (((((((((...))))).)--)))...--.((((((((...------------.....((((-....))))...))))))))..------.((((((.....))))))....... ( -21.80, z-score = -0.87, R) >droEre2.scaffold_4690 8245807 113 + 18748788 AUUCGGGAAUUAGACCAGGUCCAGCACGGAUUUCAAUUGCGAUUUCGAUUUUGAUUUGGAGUCGCUGGACCCGGAACACACGGAUUCUGAUUCGGACUGCAGAUUCCGGAUCC-- ..((.((((((...((.((((((((...((((((((...(((........)))..)))))))))))))))).))......(((((....))))).......)))))).))...-- ( -39.30, z-score = -2.38, R) >droSec1.super_8 216880 96 + 3762037 AUUCAGGAACAGGACCAGGUCCAGCUCGGAUUUCGAUUUCG------------AUUUGGAGUUGCUGGACCCGGAAUACACGGA-----AUCCGGACUGCAGAUUCCGGAUCC-- ..............((.((((((((...((((((((.....------------..)))))))))))))))).)).......(((-----.((((((........)))))))))-- ( -35.30, z-score = -2.01, R) >droSim1.chrX 13944052 96 + 17042790 AUUCAGGAACAGGACCAGGUCCAGCUCGGAUUUCGAUUUCG------------AUUUGGAGUUGCUGGACCCGGAAUACACGGA-----AACCGGACUGCAGAUUCCGGAUCC-- .....(((......((.((((((((...((((((((.....------------..)))))))))))))))).))..........-----..(((((........))))).)))-- ( -33.10, z-score = -1.63, R) >consensus AUUCAGGAACAGGACCAGGUCCAGCUCGGAUUUCGAUUUCG____________AUUUGGAGUUGCUGGACCCGGAAUACACGGA_____AUCCGGACUGCAGAUUCCGGAUCC__ .....(((......((.((((((((...((((((((...................)))))))))))))))).))................((((((........))))))))).. (-19.58 = -21.83 + 2.25)

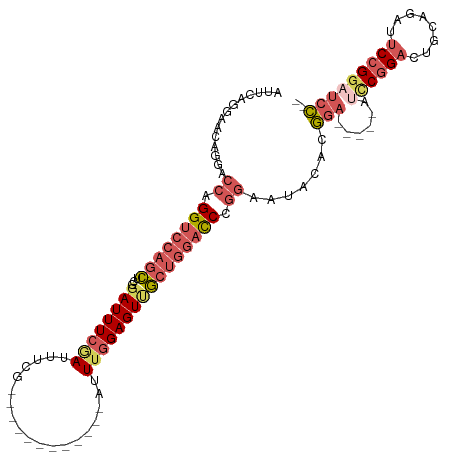
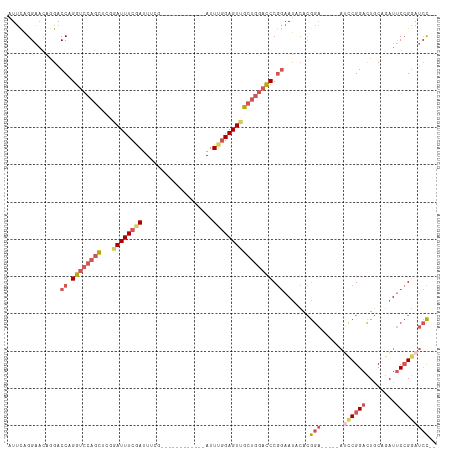
| Location | 17,916,435 – 17,916,543 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 69.78 |
| Shannon entropy | 0.50624 |
| G+C content | 0.50509 |
| Mean single sequence MFE | -35.05 |
| Consensus MFE | -19.64 |
| Energy contribution | -19.44 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.48 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.08 |
| SVM RNA-class probability | 0.997301 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 17916435 108 - 22422827 --GGAUCCGGAAUCUGCAGUCCGGAU-----UCCGUGUAUUCCGGGUCCAGCAACUCCAAAUCGAAACCGAAACCGAAAUCGAAAUCCAAGCUGGACCUGGUCUUGUUCCUAAAU --(((..(((((((((.....)))))-----))))......(((((((((((.........(((....)))...((....))........)))))))))))......)))..... ( -39.80, z-score = -4.19, R) >droAna3.scaffold_13335 1623937 92 + 3335858 GAAUGUUCCGGAUACGGAAUCCGGG------AUUUUCCAUUUUGGAAUGGGU-ACUCUGAAU------------GGGAAUGGAAAU--GGAAUG--CCAGGUCCUGAACCUGCAU ....(((((((((.....)))))))------)).((((.....)))).((((-.(((.....------------)))...(((..(--((....--)))..)))...)))).... ( -27.70, z-score = -0.72, R) >droEre2.scaffold_4690 8245807 113 - 18748788 --GGAUCCGGAAUCUGCAGUCCGAAUCAGAAUCCGUGUGUUCCGGGUCCAGCGACUCCAAAUCAAAAUCGAAAUCGCAAUUGAAAUCCGUGCUGGACCUGGUCUAAUUCCCGAAU --(((..((((.((((..........)))).))))......((((((((((((........((((...((....))...))))......))))))))))))......)))..... ( -35.24, z-score = -2.25, R) >droSec1.super_8 216880 96 - 3762037 --GGAUCCGGAAUCUGCAGUCCGGAU-----UCCGUGUAUUCCGGGUCCAGCAACUCCAAAU------------CGAAAUCGAAAUCCGAGCUGGACCUGGUCCUGUUCCUGAAU --(((..(((((((((.....)))))-----))))......(((((((((((...((.....------------.))..(((.....)))))))))))))))))........... ( -37.80, z-score = -3.49, R) >droSim1.chrX 13944052 96 - 17042790 --GGAUCCGGAAUCUGCAGUCCGGUU-----UCCGUGUAUUCCGGGUCCAGCAACUCCAAAU------------CGAAAUCGAAAUCCGAGCUGGACCUGGUCCUGUUCCUGAAU --(((.(((((........)))))..-----))).......(((((((((((...((.....------------.))..(((.....)))))))))))))).............. ( -34.70, z-score = -2.70, R) >consensus __GGAUCCGGAAUCUGCAGUCCGGAU_____UCCGUGUAUUCCGGGUCCAGCAACUCCAAAU____________CGAAAUCGAAAUCCGAGCUGGACCUGGUCCUGUUCCUGAAU ..(((((((((((..(((...(((........))))))))))))((((((((......................................)))))))).)))))........... (-19.64 = -19.44 + -0.20)

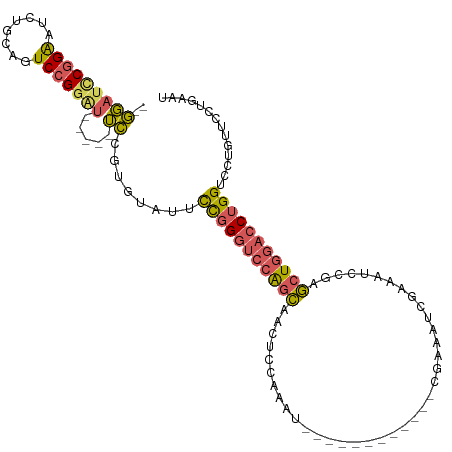
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:54:56 2011