
| Sequence ID | dm3.chrX |
|---|---|
| Location | 17,909,275 – 17,909,357 |
| Length | 82 |
| Max. P | 0.994226 |

| Location | 17,909,275 – 17,909,357 |
|---|---|
| Length | 82 |
| Sequences | 6 |
| Columns | 83 |
| Reading direction | forward |
| Mean pairwise identity | 83.48 |
| Shannon entropy | 0.31296 |
| G+C content | 0.70059 |
| Mean single sequence MFE | -43.32 |
| Consensus MFE | -33.33 |
| Energy contribution | -34.63 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.81 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.68 |
| SVM RNA-class probability | 0.994226 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
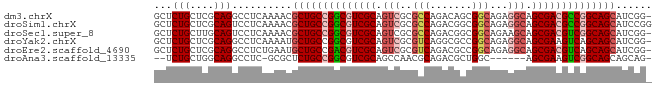
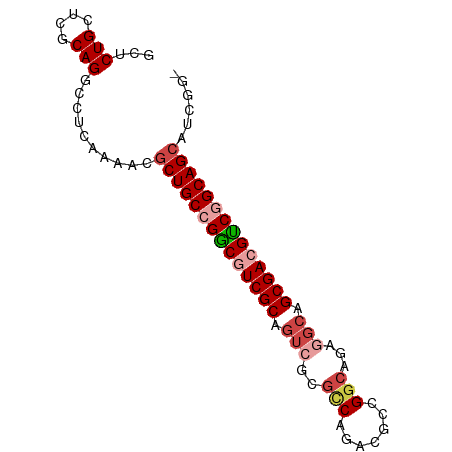
>dm3.chrX 17909275 82 + 22422827 GCUCUGCUCGCAGGCCUCAAAACGCUGCCGGCGUCGCAGUCGCGCCAGACAGCGGCAGAGGCAGCGACGCCGGCAGCAUCGG- ((.(((....)))))........((((((((((((((.(((..(((.......)))...))).)))))))))))))).....- ( -46.90, z-score = -3.51, R) >droSim1.chrX 13934960 83 + 17042790 GCUCUGCUCGCAGUCCUCAAAACGCUGCCGGCGUCGCAGUCGCGCCAGACGGCGGCAGAGGCAGCGACGCCGGCAGCAUCCGG (..(((....)))..).......((((((((((((((.(((.((((....)))).....))).))))))))))))))...... ( -45.20, z-score = -2.91, R) >droSec1.super_8 209913 82 + 3762037 GCUCUGCUUGCAGUCCUCAAAACGCUGCCGGCGUCGCAGUCGCGCCAGACGGCGGCAGAAGCAGCGACGUCGGCAGCAUCGG- (..(((....)))..).......((((((((((((((.....((((....))))((....)).)))))))))))))).....- ( -42.30, z-score = -2.80, R) >droYak2.chrX 16529564 82 + 21770863 GCUCUGCUCGCAGGCCUCAAAAUGCUGCCGGCGUCGCAGUCGCGUCAGGCGCCGGCAGAGGCAGCGAAGUCAGCAGCAUCGG- (((((((((((..(((((.......((((((((((((....)))....)))))))))))))).))).)).))).))).....- ( -41.01, z-score = -2.48, R) >droEre2.scaffold_4690 8236996 82 + 18748788 GCUCUGCUCGCAGGCCUCUGAAUGCUGCCGACGUCGCAGUCGCGUCAGACGCCGGCAGAGGCAGCGACGUCAGCAGCAUCGG- ((.(((....)))))..((((.((((((.((((((((.(((......)))(((......))).)))))))).))))))))))- ( -44.20, z-score = -3.04, R) >droAna3.scaffold_13335 1617098 73 - 3335858 --UCUGCUGGCAGGCCUC-GCGCUCUGCCGGCGUCGCAGCCAACGCAGACGCUGGC------AGCGAAGUCGGCAGCAGCAG- --.((((((.(.(((.((-((....((((((((((((.......)).)))))))))------))))).)))).))))))...- ( -40.30, z-score = -2.11, R) >consensus GCUCUGCUCGCAGGCCUCAAAACGCUGCCGGCGUCGCAGUCGCGCCAGACGCCGGCAGAGGCAGCGACGUCGGCAGCAUCGG_ ...(((....)))..........((((((((((((((.(((..(((.......)))...))).))))))))))))))...... (-33.33 = -34.63 + 1.31)
| Location | 17,909,275 – 17,909,357 |
|---|---|
| Length | 82 |
| Sequences | 6 |
| Columns | 83 |
| Reading direction | reverse |
| Mean pairwise identity | 83.48 |
| Shannon entropy | 0.31296 |
| G+C content | 0.70059 |
| Mean single sequence MFE | -42.13 |
| Consensus MFE | -28.77 |
| Energy contribution | -29.55 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.53 |
| SVM RNA-class probability | 0.947418 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
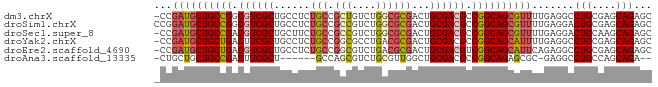
>dm3.chrX 17909275 82 - 22422827 -CCGAUGCUGCCGGCGUCGCUGCCUCUGCCGCUGUCUGGCGCGACUGCGACGCCGGCAGCGUUUUGAGGCCUGCGAGCAGAGC -..(((((((((((((((((....(((((((.....))))).))..))))))))))))))))).....(((((....))).)) ( -45.90, z-score = -2.61, R) >droSim1.chrX 13934960 83 - 17042790 CCGGAUGCUGCCGGCGUCGCUGCCUCUGCCGCCGUCUGGCGCGACUGCGACGCCGGCAGCGUUUUGAGGACUGCGAGCAGAGC ((.(((((((((((((((((....((.((.(((....)))))))..)))))))))))))))))....)).(((....)))... ( -46.60, z-score = -2.70, R) >droSec1.super_8 209913 82 - 3762037 -CCGAUGCUGCCGACGUCGCUGCUUCUGCCGCCGUCUGGCGCGACUGCGACGCCGGCAGCGUUUUGAGGACUGCAAGCAGAGC -((((((((((((.((((((.((....))((((....)))).....)))))).))))))))))....)).(((....)))... ( -38.60, z-score = -1.60, R) >droYak2.chrX 16529564 82 - 21770863 -CCGAUGCUGCUGACUUCGCUGCCUCUGCCGGCGCCUGACGCGACUGCGACGCCGGCAGCAUUUUGAGGCCUGCGAGCAGAGC -.....(((.(((.((.(((.(((((((((((((.....(((....))).)))))))).......)))))..))))))))))) ( -36.81, z-score = -2.02, R) >droEre2.scaffold_4690 8236996 82 - 18748788 -CCGAUGCUGCUGACGUCGCUGCCUCUGCCGGCGUCUGACGCGACUGCGACGUCGGCAGCAUUCAGAGGCCUGCGAGCAGAGC -..(((((((((((((((((.(((......)))(((......))).))))))))))))))))).....(((((....))).)) ( -43.30, z-score = -2.67, R) >droAna3.scaffold_13335 1617098 73 + 3335858 -CUGCUGCUGCCGACUUCGCU------GCCAGCGUCUGCGUUGGCUGCGACGCCGGCAGAGCGC-GAGGCCUGCCAGCAGA-- -((((((((((((.(.((((.------(((((((....))))))).)))).).))))))...((-(.....))))))))).-- ( -41.60, z-score = -2.30, R) >consensus _CCGAUGCUGCCGACGUCGCUGCCUCUGCCGCCGUCUGGCGCGACUGCGACGCCGGCAGCGUUUUGAGGCCUGCGAGCAGAGC ...((((((((((.((((((......(((.(((....))))))...)))))).)))))))))).......(((....)))... (-28.77 = -29.55 + 0.78)
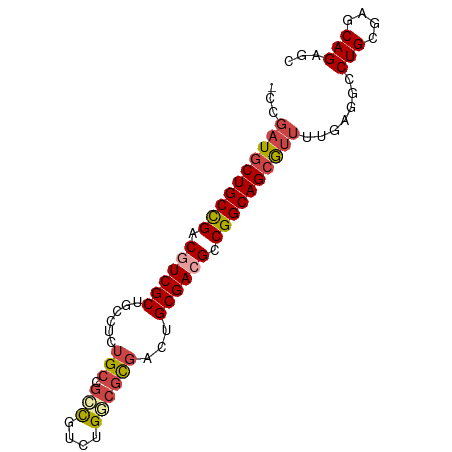
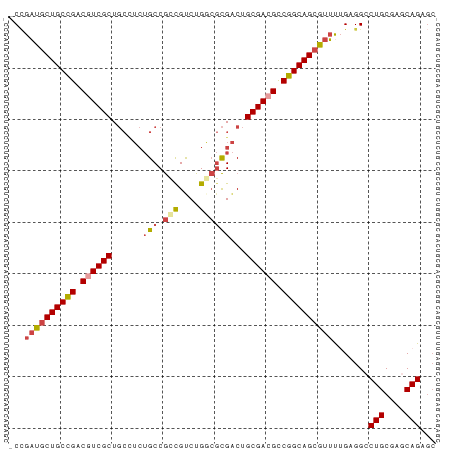
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:54:51 2011