
| Sequence ID | dm3.chrX |
|---|---|
| Location | 17,906,487 – 17,906,591 |
| Length | 104 |
| Max. P | 0.995379 |

| Location | 17,906,487 – 17,906,591 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 59.12 |
| Shannon entropy | 0.75195 |
| G+C content | 0.45569 |
| Mean single sequence MFE | -25.40 |
| Consensus MFE | -12.66 |
| Energy contribution | -14.08 |
| Covariance contribution | 1.42 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.80 |
| SVM RNA-class probability | 0.995379 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
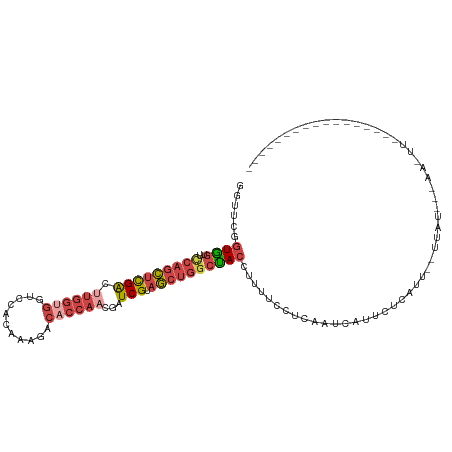
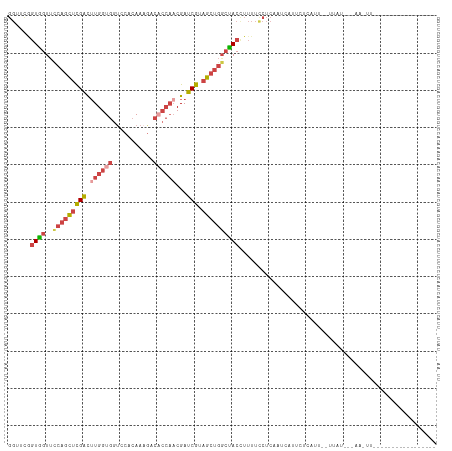
>dm3.chrX 17906487 104 + 22422827 GGUUCGGUGGUUCCAGCUCGACUUGGUGGUCCACAAAGACACCAACGAUCGUAGCUGUCUACCUUUUCCUCAAACAUUAUCAUU--UUUCAAAAAUUUUUUGUGAU--------- ((...(((((...(((((((((((((((.((......)))))))).).))).))))).)))))....)).........(((((.--...............)))))--------- ( -26.39, z-score = -2.13, R) >droSim1.chrX 13932183 88 + 17042790 GGUUCAGUAGUUCCAGCUCGACUUGGUGGUCCACAAAGACACCAACGAUCGUAGCUGGCUACCUUUUCCUCAAUCAAUCUCAUU--UGAU------------------------- ((....((((..((((((((((((((((.((......)))))))).).))).)))))))))).....))...(((((......)--))))------------------------- ( -27.20, z-score = -3.54, R) >droSec1.super_8 207118 88 + 3762037 GGUUCAGUAGUUUCAGCUCGACUUGGUGGUCCACAAAGACACCAACGAUCGUAGCUGGCUACCUUUUUCUCAAUGAAUCUCAUU--UGAU------------------------- (((((((((((..(((((((((((((((.((......)))))))).).))).))))))))))...........)))))).....--....------------------------- ( -25.60, z-score = -2.56, R) >droYak2.chrX 16526785 102 + 21770863 GGCUCGGUGGUUUCAGCUCGACUUGGUGGUUCACAA---------CGAUCGCAGCUGGCUACUUUUUCCUCAUCCAUACCCAUUGCUUAUCUCAACUUAUACUGUUUUGUU---- ((...((((((..((((((((.(((.((.....)).---------)))))).)))))))))))....))..........................................---- ( -18.60, z-score = 0.27, R) >droEre2.scaffold_4690 8234284 115 + 18748788 GGCAUGGUGGUUCCAGCUCGACUUGGUGGCUCACAAAGACACCAACGAUCGUAGCUGGCUACUUUUACCUCACCCAUUCUCGCGGCACUUUAAAAUUGUCACGAUUUUUAGAGAU ((...((((((.((((((((((((((((.((.....)).)))))).).))).))))))..)))...)))....))....(((.((((.........)))).)))........... ( -31.80, z-score = -1.74, R) >droAna3.scaffold_13335 1614212 91 - 3335858 -------UGGUCUCUCUAUGGUAUGGUGGUUCCAAAAGCCUCCAACGAUCGGCAAUUGACACAUUUCUCUGGCUCUUUUCGAAGGGGUAGUGGGAAUU----------------- -------..(((......((((.(((.((((.....)))).)))...))))......)))..(((((((((.(((((....))))).))).)))))).----------------- ( -22.80, z-score = 0.15, R) >consensus GGUUCGGUGGUUCCAGCUCGACUUGGUGGUCCACAAAGACACCAACGAUCGUAGCUGGCUACCUUUUCCUCAAUCAUUCUCAUU__UUAU___AA_UU_________________ ......((((..(((((((((.((((((...........))))))...))).))))))))))..................................................... (-12.66 = -14.08 + 1.42)

| Location | 17,906,487 – 17,906,591 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 59.12 |
| Shannon entropy | 0.75195 |
| G+C content | 0.45569 |
| Mean single sequence MFE | -23.94 |
| Consensus MFE | -10.62 |
| Energy contribution | -11.65 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.08 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.918840 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
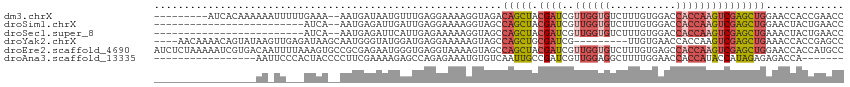
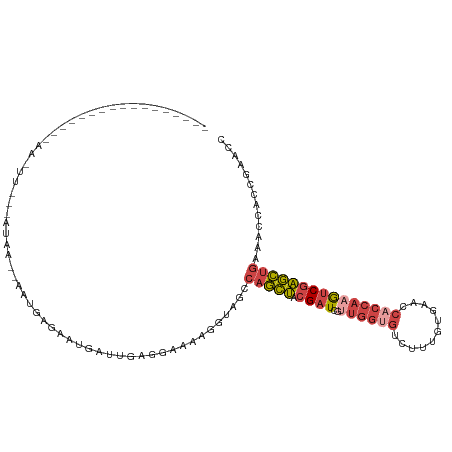
>dm3.chrX 17906487 104 - 22422827 ---------AUCACAAAAAAUUUUUGAAA--AAUGAUAAUGUUUGAGGAAAAGGUAGACAGCUACGAUCGUUGGUGUCUUUGUGGACCACCAAGUCGAGCUGGAACCACCGAACC ---------.((........(((((.(((--.((....)).))).)))))..(((...(((((.((((..(((((((((....))).)))))))))))))))..)))...))... ( -26.60, z-score = -1.57, R) >droSim1.chrX 13932183 88 - 17042790 -------------------------AUCA--AAUGAGAUUGAUUGAGGAAAAGGUAGCCAGCUACGAUCGUUGGUGUCUUUGUGGACCACCAAGUCGAGCUGGAACUACUGAACC -------------------------((((--(......)))))...((....(((((((((((.((((..(((((((((....))).))))))))))))))))..)))))...)) ( -29.80, z-score = -3.26, R) >droSec1.super_8 207118 88 - 3762037 -------------------------AUCA--AAUGAGAUUCAUUGAGAAAAAGGUAGCCAGCUACGAUCGUUGGUGUCUUUGUGGACCACCAAGUCGAGCUGAAACUACUGAACC -------------------------.(((--(.((.....))))))......(((((.(((((.((((..(((((((((....))).)))))))))))))))...)))))..... ( -26.80, z-score = -2.86, R) >droYak2.chrX 16526785 102 - 21770863 ----AACAAAACAGUAUAAGUUGAGAUAAGCAAUGGGUAUGGAUGAGGAAAAAGUAGCCAGCUGCGAUCG---------UUGUGAACCACCAAGUCGAGCUGAAACCACCGAGCC ----...............(((......)))....(((.(((.((.((.........))((((.((((.(---------(.(....).))...)))))))).....))))).))) ( -15.60, z-score = 1.24, R) >droEre2.scaffold_4690 8234284 115 - 18748788 AUCUCUAAAAAUCGUGACAAUUUUAAAGUGCCGCGAGAAUGGGUGAGGUAAAAGUAGCCAGCUACGAUCGUUGGUGUCUUUGUGAGCCACCAAGUCGAGCUGGAACCACCAUGCC .((.((.....(((((.((.........)).))))).....)).))((((.......((((((.((((..((((((.(((...))).))))))))))))))))........)))) ( -33.36, z-score = -1.62, R) >droAna3.scaffold_13335 1614212 91 + 3335858 -----------------AAUUCCCACUACCCCUUCGAAAAGAGCCAGAGAAAUGUGUCAAUUGCCGAUCGUUGGAGGCUUUUGGAACCACCAUACCAUAGAGAGACCA------- -----------------...((.(.(((..........(((((((.....((((.(((.......)))))))...)))))))((.....))......))).).))...------- ( -11.50, z-score = 1.60, R) >consensus _________________AA_UU___AUAA__AAUGAGAAUGAUUGAGGAAAAGGUAGCCAGCUACGAUCGUUGGUGUCUUUGUGAACCACCAAGUCGAGCUGAAACCACCGAACC ..........................................................(((((.((((..((((((...........)))))))))))))))............. (-10.62 = -11.65 + 1.03)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:54:50 2011