| Sequence ID | dm3.chrX |
|---|---|
| Location | 17,883,603 – 17,883,659 |
| Length | 56 |
| Max. P | 0.984056 |

| Location | 17,883,603 – 17,883,659 |
|---|---|
| Length | 56 |
| Sequences | 9 |
| Columns | 70 |
| Reading direction | forward |
| Mean pairwise identity | 66.40 |
| Shannon entropy | 0.63458 |
| G+C content | 0.49919 |
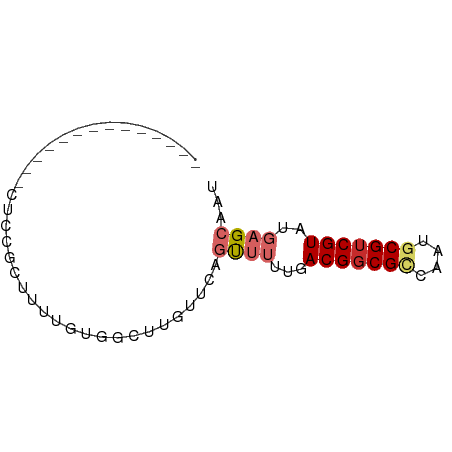
| Mean single sequence MFE | -17.04 |
| Consensus MFE | -9.66 |
| Energy contribution | -10.37 |
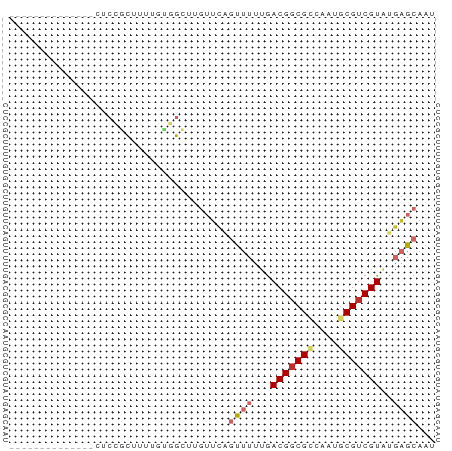
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.15 |
| SVM RNA-class probability | 0.984056 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
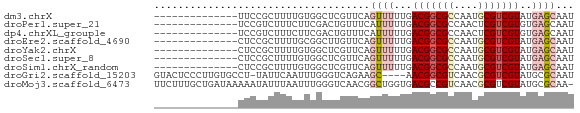
>dm3.chrX 17883603 56 + 22422827 --------------UUCCGCUUUUGUGGCUCGUUCAGUUUUUGACGGCGCCAAUGCGUCGUAUGAGCAAU --------------..((((....))))...(((((.......(((((((....))))))).)))))... ( -18.80, z-score = -2.08, R) >droPer1.super_21 136117 56 + 1645244 --------------UCCGUCUUUCUUCGACUGUUUCAUUUUUGACGGCGCCAACUCGUCGUGUGAGCAAU --------------...(((.......)))((((.(((...((((((.......)))))).))))))).. ( -10.80, z-score = -0.05, R) >dp4.chrXL_group1e 5227629 56 + 12523060 --------------UCCGUCUUUCUUCGACUGUUUCAUUUUUGACGGCGCCAACUCGUCGUGUGAGCAAU --------------...(((.......)))((((.(((...((((((.......)))))).))))))).. ( -10.80, z-score = -0.05, R) >droEre2.scaffold_4690 8211423 56 + 18748788 --------------CUCCGCUUUUGCGGCUUGUUCAGUUUUUGACGGCGCCAAUGCGUCGUAUGAGCAAU --------------..((((....)))).(((((((.......(((((((....))))))).))))))). ( -21.90, z-score = -3.00, R) >droYak2.chrX 16502514 56 + 21770863 --------------CUCCGCUUUUGUGGCUCGUUCAGUUUUUGACGGCGCCAAUGCGUCGUAUGAGCAAU --------------..((((....))))...(((((.......(((((((....))))))).)))))... ( -18.80, z-score = -2.07, R) >droSec1.super_8 184212 56 + 3762037 --------------CUCCGCUUUUGUGGCUCGUUCAGUUUUUGACGGCGCCAAUGCGUCGUAUGAGCAAU --------------..((((....))))...(((((.......(((((((....))))))).)))))... ( -18.80, z-score = -2.07, R) >droSim1.chrX_random 4583825 56 + 5698898 --------------CUCCGCUUUUGUGGCUCGUUCAGUUUUUGACGGCGCCAAUGCGUCGUAUGAGCAAU --------------..((((....))))...(((((.......(((((((....))))))).)))))... ( -18.80, z-score = -2.07, R) >droGri2.scaffold_15203 1653553 65 + 11997470 GUACUCCCUUGUGCCU-UAUUCAAUUUGGGUCAGAAGC----AACGGCGUCAACGCGUCGUAUGCGCAAU ((((......))))((-.((((.....)))).))..((----((((((((....))))))).)))..... ( -17.00, z-score = -0.80, R) >droMoj3.scaffold_6473 2339504 69 - 16943266 UUCUUUGCUGAUAAAAAUAUUUAAUUUGGGUCAACGGCUGGUGACGCCGUCAACGCGUCGUAUGCGCAA- ........((((..((((.....))))..))))(((((.(....))))))....((((.....))))..- ( -17.70, z-score = -0.67, R) >consensus ______________CUCCGCUUUUGUGGCUUGUUCAGUUUUUGACGGCGCCAAUGCGUCGUAUGAGCAAU ....................................((((...(((((((....)))))))..))))... ( -9.66 = -10.37 + 0.70)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:54:45 2011