| Sequence ID | dm3.chrX |
|---|---|
| Location | 17,877,369 – 17,877,456 |
| Length | 87 |
| Max. P | 0.917707 |

| Location | 17,877,369 – 17,877,456 |
|---|---|
| Length | 87 |
| Sequences | 7 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 66.38 |
| Shannon entropy | 0.64526 |
| G+C content | 0.45871 |
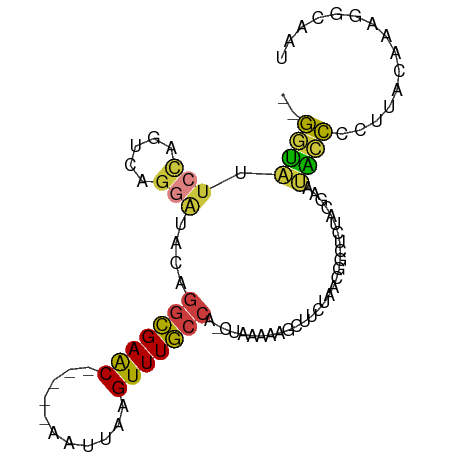
| Mean single sequence MFE | -22.81 |
| Consensus MFE | -9.01 |
| Energy contribution | -8.40 |
| Covariance contribution | -0.61 |
| Combinations/Pair | 1.64 |
| Mean z-score | -1.75 |
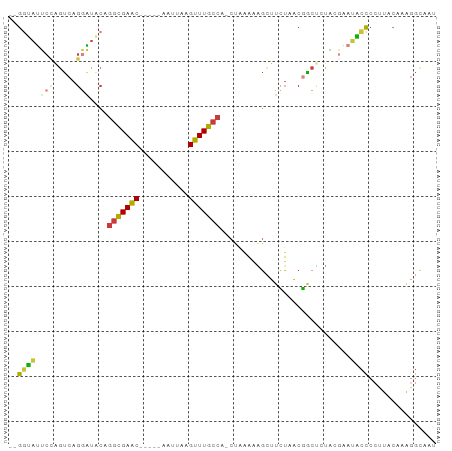
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.917707 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
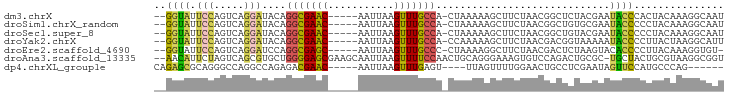
>dm3.chrX 17877369 87 + 22422827 --GGUAUUCCAGUCAGGAUACAGGCGAAC-----AAUUAAGUUUGCCA-CUAAAAAGCUUCUAACGGCUCUACGAAUACCCACUACAAAGGCAAU --(((((((.............(((((((-----......))))))).-......((((......))))....)))))))............... ( -21.20, z-score = -2.72, R) >droSim1.chrX_random 4576657 87 + 5698898 --GGUAUUCCAGUCAGGAUACAGGCGAAC-----AAUUAAGUUUGCCA-CUAAAAAGCUUCUAACGGCUGUGCGAAUACCCCCUACAAAGGCAAU --((((((((((((.(....).(((((((-----......))))))).-................)))))...))))))).(((....))).... ( -24.60, z-score = -2.49, R) >droSec1.super_8 178126 87 + 3762037 --GGUAUUCCAGUCAGGAUACAGGCGAAC-----AAUUAAGUUUGCCA-CUAAAAAGCUUCUAACGGCUGUACGAAUACCCCCUACAAAGGCAAU --((((((((((((.(....).(((((((-----......))))))).-................)))))...))))))).(((....))).... ( -24.60, z-score = -3.06, R) >droYak2.chrX 16496209 87 + 21770863 --GGUAUUCCAGUCAGGAUACAGGCGAAC-----AAUUAAGUUUGCCA-CCAAAAAGCUUCUAACGACGGUAAAAAUACCCCUUACUAAGGCAUU --(((..(((.....)))....(((((((-----......))))))))-)).....((((........((((....))))........))))... ( -21.29, z-score = -2.46, R) >droEre2.scaffold_4690 8205581 86 + 18748788 --GGUAUUCCAGUCAGGAUCCAGGCGAGC-----AAUUAAGUUUGCCC-CUAAAAGGCUUCUAACGACUCUAAGUACACCCCUUACAAAGGUGU- --((...(((.....))).)).(((((((-----......))))))).-......((..((....))..))....(((((.........)))))- ( -19.50, z-score = -0.55, R) >droAna3.scaffold_13335 1580170 92 - 3335858 --AACAUUCUAGUCAGCGUGCUGGGGAGCGAAGCAAUUAAGUUUUCCAACUGCAGGGAAAGUGUCCAGACUGCGC-UGCUACUGCGUAAGGCGGU --.......(((.((((....(((..(((...........)))..)))...(((((((.....)))...))))))-)))))((((.....)))). ( -26.60, z-score = 0.59, R) >dp4.chrXL_group1e 5219718 80 + 12523060 CAGAGCGCAGGGCCAGGCCAGAGACGAAC-----AAUUAAGUUUGAGU----UUAGUUUUGGAACUGCCUCGAAUAGUUCCAUGCCCAG------ ..(((.((((..((((((.(((..(((((-----......)))))..)----)).).)))))..)))))))..................------ ( -21.90, z-score = -1.53, R) >consensus __GGUAUUCCAGUCAGGAUACAGGCGAAC_____AAUUAAGUUUGCCA_CUAAAAAGCUUCUAACGGCUCUACGAAUACCCCUUACAAAGGCAAU ..((((.(((.....)))....(((((((...........))))))).............................))))............... ( -9.01 = -8.40 + -0.61)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:54:41 2011