| Sequence ID | dm3.chrX |
|---|---|
| Location | 17,875,359 – 17,875,453 |
| Length | 94 |
| Max. P | 0.923532 |

| Location | 17,875,359 – 17,875,453 |
|---|---|
| Length | 94 |
| Sequences | 8 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 79.13 |
| Shannon entropy | 0.36796 |
| G+C content | 0.40203 |
| Mean single sequence MFE | -25.10 |
| Consensus MFE | -11.39 |
| Energy contribution | -12.60 |
| Covariance contribution | 1.21 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.80 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.923532 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
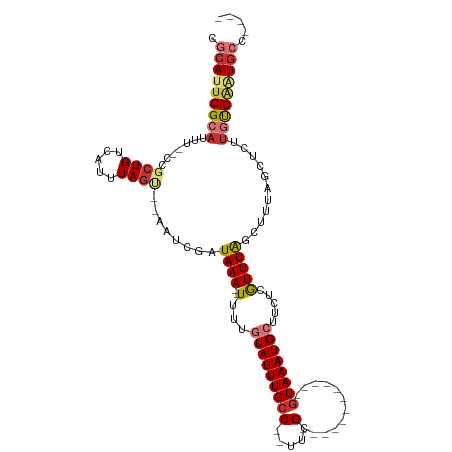
>dm3.chrX 17875359 94 - 22422827 CGCAUUGGCAUUU--CCGCUGUCAUUUAGU--AAUCGAUAAA--UUUUGUAUUUGCC--UUCG------------GUAAAUGCUUCUCGUUUGGCUUUAGCUCUUGUCAAUGCC---- .(((((((((...--..((((......(((--...(((....--....(((((((((--...)------------))))))))...)))....))).))))...))))))))).---- ( -25.50, z-score = -3.41, R) >droWil1.scaffold_181150 1845791 117 + 4952429 -UCAUUGGCAUUUUCCUGCUGUCAUUUAAGUAAAUCGAUAAAAUUGUUGUAUUUGGCUUUGGGCUGGGUAACAGUGUAAAUGCUUUGCAUUUAGAUUUUAGUUGCAUCUGUAUUCGGC -.....((((......)))).........((((.....(((((((..((((...(((((((.((((.....)))).)))).))).))))....))))))).))))............. ( -23.20, z-score = 0.41, R) >droPer1.super_21 125916 92 - 1645244 CGCAUUGGCAUUU--CCGCUGUCAUUUAGG--AAUCGAUAAA--UGUAGUAUUUGCC--UUUG------------GUAAAUGAUUUUCGUUUAG--UUGGCUUUUGUCGAUGCC---- .(((((((((...--..((((.((((((..--......))))--))))))....(((--...(------------.((((((.....)))))).--).)))...))))))))).---- ( -27.50, z-score = -3.21, R) >dp4.chrXL_group1e 5217648 92 - 12523060 CGCAUUGGCAUUU--CCGCUGUCAUUUAGU--AAUCGAUAAA--UGUAGUAUUUGCC--UUUG------------GUAAAUGAUUUUCGUUUAG--UUGGCUUUUGUCGCUGCC---- .(((.(((((...--..((((.((((((..--......))))--))))))....(((--...(------------.((((((.....)))))).--).)))...))))).))).---- ( -23.40, z-score = -2.16, R) >droAna3.scaffold_13335 1578829 95 + 3335858 CUCAUUGGCAUUU--CCGCUGUCAUUUAGU--AAUCGAUAAA--UUUUGUAUUUGCCUUUUCG------------GUAAAUGCUUUUCGUUUAGC-CCUGCCACUACCACUGCC---- .....(((((...--..((((.....))))--......((((--(...(((((((((.....)------------)))))))).....)))))..-..)))))...........---- ( -19.10, z-score = -2.09, R) >droEre2.scaffold_4690 8203660 93 - 18748788 CGCAUUGGCAUUU--CCGCUGUCAUUUAGC--AAUCGAUAAA--UUUUGUAUUUGCC--UUCG------------GUAAAUGC-UCUCGUUUGGCUUCAGCUCUUGUCAAUGCC---- .(((((((((...--..((((......(((--...(((....--....(((((((((--...)------------))))))))-..)))....))).))))...))))))))).---- ( -28.50, z-score = -3.92, R) >droYak2.chrX 16494201 94 - 21770863 CGCAUUGGCAUUU--CCGCUGUCAUUUAGU--AAUCGAUAAA--UUUUGUAUUUGCC--CUCG------------GUAAAUGCUUCUCAUUUGCCUUCAGCCGUUGUCAAUGCC---- .(((((((((...--(.((((.......((--((.(((....--..)))...)))).--...(------------(((((((.....))))))))..)))).).))))))))).---- ( -28.10, z-score = -4.57, R) >droSec1.super_8 176321 94 - 3762037 CGCAUUGGCAUUU--CCGCUGUCAUUUAGU--AAUCGAUAAA--UUUUGUAUUUGCC--UUCG------------GUAAAUGCUUCUCGUUUGGCUUUAGCUCUUGUCAAUGCC---- .(((((((((...--..((((......(((--...(((....--....(((((((((--...)------------))))))))...)))....))).))))...))))))))).---- ( -25.50, z-score = -3.41, R) >consensus CGCAUUGGCAUUU__CCGCUGUCAUUUAGU__AAUCGAUAAA__UUUUGUAUUUGCC__UUCG____________GUAAAUGCUUCUCGUUUAGCUUUAGCUCUUGUCAAUGCC____ .(((((((((.......((((.....))))........((((......((((((((...................))))))))......))))...........)))))))))..... (-11.39 = -12.60 + 1.21)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:54:40 2011