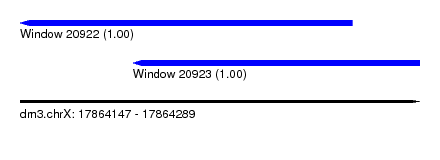
| Sequence ID | dm3.chrX |
|---|---|
| Location | 17,864,147 – 17,864,289 |
| Length | 142 |
| Max. P | 0.998530 |

| Location | 17,864,147 – 17,864,265 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.25 |
| Shannon entropy | 0.40444 |
| G+C content | 0.45018 |
| Mean single sequence MFE | -31.35 |
| Consensus MFE | -22.92 |
| Energy contribution | -22.95 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.79 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.09 |
| SVM RNA-class probability | 0.997356 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 17864147 118 - 22422827 --UCGGAAAAUAUGCAUUUUCCGCGUUUUUCCGACCCAACCGACCGACCCCUCGCUGGUAAUAAAUUUGGCUAGCAGCUUAACAAGGGAGCUCAACAGCAAUGAACAAAAACAAAAAAUA --((((((((.((((.......))))))))))))(((........((....))((((((..........))))))..........))).(((....)))..................... ( -25.70, z-score = -1.16, R) >droSim1.chrX 13913081 114 - 17042790 --UCGGAAAAUAUGCAUUUUCCGCGUUUUUCCGCCACA----CCCGACCCCUCGCUGGCCAUAAAUUUGGCUAGCAGCUUAACAAGGGAGCUGAACAGCAAUGAACAAAAACAAAAAAAA --.(((((((.((((.......))))))))))).....----(((((....))((((((((......))))))))..........))).(((....)))..................... ( -32.80, z-score = -3.09, R) >droSec1.super_8 165525 114 - 3762037 --UCGGAAAAUACGCAUUAUCCGCGUUUUUCCGCCACA----CCCGACCCCUCGCUGGCCAUAAAUUUGGCUAGCAGCUUAACAAGGGAGCUGAACAGCAAUGAACAAAAACAAAAAAAA --.(((((((.((((.......))))))))))).....----(((((....))((((((((......))))))))..........))).(((....)))..................... ( -34.60, z-score = -3.99, R) >droYak2.chrX 16483185 107 - 21770863 --UAGGAAAAUAUGCAUUUUCCGCGUUUUUCCGCCCCA----CCCGACCCCUCGCUGGCCAUAAAUUUGGCUAGCAGCUUAACAAGGGAGCUGAACAGCAAUGAAAAAAAAAA------- --..(((((((....)))))))..(((.(((.(((((.----...((....))((((((((......))))))))..........))).)).))).)))..............------- ( -33.30, z-score = -3.17, R) >droEre2.scaffold_4690 8189792 106 - 18748788 --UCGGAAAAUAUGCAUUUUCCGCGCCUUUGCGCUCCA----CCCGACCCCUUGCUGGCCAUAAAUUUGGCCAGCAGCUUAACAA-GAAGCUGAAUAGCAACGAACCCAAAGA------- --..(((((((....)))))))((((....))))....----..((.......((((((((......))))))))(((((.....-.))))).........))..........------- ( -34.20, z-score = -3.96, R) >droAna3.scaffold_13335 1566871 108 + 3335858 CUGCCAAAAAUAUGCAUUUUUCG------GCAGCUCUA------UGGGGAAUCGUUGGCCAUAAAUUUGGUUAGCAGCUUAACAAGUGAGCCGAAGAGGACCAAAAAAAUCCACAAAAAU (((((((((((....)))))).)------))))((((.------((.......((((((((......)))))))).(((((.....))))))).))))...................... ( -27.50, z-score = -1.37, R) >consensus __UCGGAAAAUAUGCAUUUUCCGCGUUUUUCCGCCCCA____CCCGACCCCUCGCUGGCCAUAAAUUUGGCUAGCAGCUUAACAAGGGAGCUGAACAGCAAUGAACAAAAACAAAAAAAA ....(((((((....)))))))((.............................((((((((......))))))))(((((.......))))).....))..................... (-22.92 = -22.95 + 0.03)

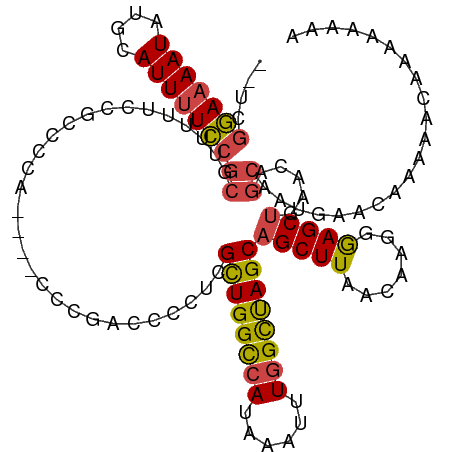
| Location | 17,864,187 – 17,864,289 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 74.76 |
| Shannon entropy | 0.45983 |
| G+C content | 0.55163 |
| Mean single sequence MFE | -32.77 |
| Consensus MFE | -19.09 |
| Energy contribution | -20.04 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.32 |
| Mean z-score | -3.09 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.39 |
| SVM RNA-class probability | 0.998530 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
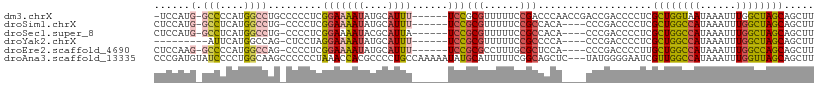
>dm3.chrX 17864187 102 - 22422827 -UCCAUG-GCCCCAUGGCCUGCCCCCUCGGAAAAUAUGCAUUU------UCCGCGUUUUUCCGACCCAACCGACCGACCCCUCGCUGGUAAUAAAUUUGGCUAGCAGCUU -.(((((-....))))).((((..((((((((((.((((....------...))))))))))))....((((..(((....))).)))).........))...))))... ( -28.30, z-score = -1.94, R) >droSim1.chrX 13913121 98 - 17042790 CUCCAUG-GCCUCAUGGCCUG-CCCCUCGGAAAAUAUGCAUUU------UCCGCGUUUUUCCGCCACA----CCCGACCCCUCGCUGGCCAUAAAUUUGGCUAGCAGCUU ......(-(((....)))).(-(....(((((((.((((....------...))))))))))).....----...........((((((((......)))))))).)).. ( -33.90, z-score = -3.72, R) >droSec1.super_8 165565 98 - 3762037 CUCCAUG-GCCUCAUGGCCUG-CCCCUCGGAAAAUACGCAUUA------UCCGCGUUUUUCCGCCACA----CCCGACCCCUCGCUGGCCAUAAAUUUGGCUAGCAGCUU ......(-(((....)))).(-(....(((((((.((((....------...))))))))))).....----...........((((((((......)))))))).)).. ( -35.70, z-score = -4.43, R) >droYak2.chrX 16483218 90 - 21770863 ---------AUUCAUGGCCAG-CUCCUAGGAAAAUAUGCAUUU------UCCGCGUUUUUCCGCCCCA----CCCGACCCCUCGCUGGCCAUAAAUUUGGCUAGCAGCUU ---------....((((((((-(.....(((((((....))))------)))(((......)))....----...........)))))))))......(((.....))). ( -28.20, z-score = -3.00, R) >droEre2.scaffold_4690 8189824 98 - 18748788 CUCCAAG-GCCCCAUGGCCAG-CCCCUCGGAAAAUAUGCAUUU------UCCGCGCCUUUGCGCUCCA----CCCGACCCCUUGCUGGCCAUAAAUUUGGCCAGCAGCUU ......(-(((....))))((-(...(((((((((....))))------)))((((....))))....----...)).....(((((((((......)))))))))))). ( -38.90, z-score = -4.34, R) >droAna3.scaffold_13335 1566911 107 + 3335858 CCCGAUGUAUCCCCUGGCAAGCCCCCCUAAACCACGCCCCUGCCAAAAAUAUGCAUUUUUCGGCAGCUC---UAUGGGGAAUCGUUGGCCAUAAAUUUGGUUAGCAGCUU ...(((...(((((.(((.................))).(((((((((((....)))))).)))))...---...))))))))((((((((......))))))))..... ( -31.63, z-score = -1.12, R) >consensus CUCCAUG_GCCCCAUGGCCAG_CCCCUCGGAAAAUAUGCAUUU______UCCGCGUUUUUCCGCCCCA____CCCGACCCCUCGCUGGCCAUAAAUUUGGCUAGCAGCUU ...............(((.........(((((((.((((.............)))))))))))....................((((((((......)))))))).))). (-19.09 = -20.04 + 0.95)
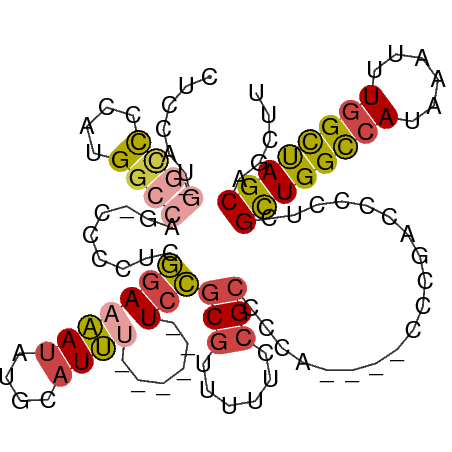
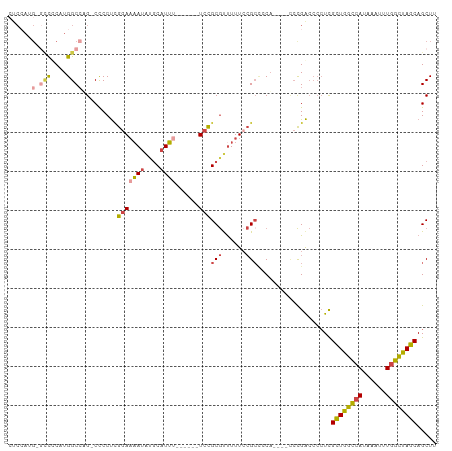
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:54:37 2011