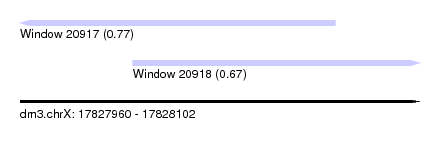
| Sequence ID | dm3.chrX |
|---|---|
| Location | 17,827,960 – 17,828,102 |
| Length | 142 |
| Max. P | 0.771704 |

| Location | 17,827,960 – 17,828,072 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 72.30 |
| Shannon entropy | 0.49697 |
| G+C content | 0.51723 |
| Mean single sequence MFE | -29.46 |
| Consensus MFE | -16.00 |
| Energy contribution | -16.24 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.771704 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
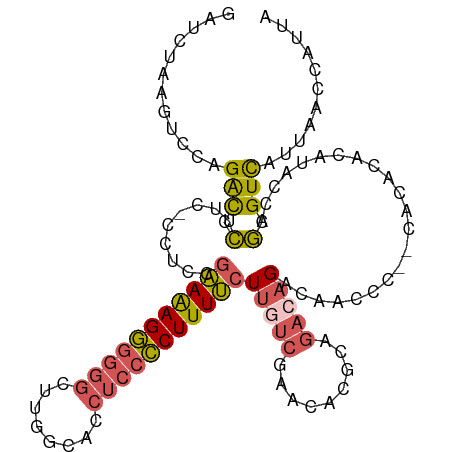
>dm3.chrX 17827960 112 - 22422827 GAUCUAAGUCCAGAUCCUUC-CCUACGAAAAGGGGGGCUUGGCACCUCCCCUUUUCUUGUCGAACACGCAGACAGACAACCCUACACACACACACCCGAGUCAUUUACCAUUA (((((......)))))....-.....(((((((((((........))))))))))).((((.........))))(((......................)))........... ( -24.65, z-score = -1.30, R) >droSec1.super_8 129776 110 - 3762037 GAUCUAAGUCCAGACCCUUC-CCUCCGAAAAGGGGGGCUUGGCACCUCCCCUUUUCUUUUCGGACACACAGACAGACCACCC--CACACACACACUCGAGUCAUAUACCAUUA .......((((.((....))-.....(((((((((((........))))))))))).....)))).....(((.((......--...........))..)))........... ( -23.53, z-score = -0.95, R) >droYak2.chrX 16446650 112 - 21770863 GAUCUAAGUCCAGACCCUUC-GUUUCGAAGAGGGGGGCUUGGCACCUCCCCUUUUCUUGUCGAACACGCAGACAGACAACCCCAUACACACAUACCCGAGUCAUUAACCAUUA ............(((...((-(....(((((((((((........)))))))))))(((((.........))))).....................))))))........... ( -24.80, z-score = -0.69, R) >droEre2.scaffold_4690 8153388 109 - 18748788 GAUCUAAGUCCAGACCCUUC-GCUCUGAAAAGGGGGGCUUGGCACCUCCCCUUUUCUUGUCGAACACGCAGACAGACAACC---CACACACAUACCCGAGUCAUAAACCAUUA ....................-((((.(((((((((((........)))))))))))(((((.........)))))......---.............))))............ ( -24.30, z-score = -0.66, R) >droVir3.scaffold_12928 4827822 110 + 7717345 UAUGCAAGUAUGGGUUAGUCAAGGCCUGAGAGCAGAGCUGUGCUUGAGAGCUUUCCUGUGAGCUCAC-CAGACGGGCGUGA--GCGCAGCUCUGCCUUAGUUGUUGGCAAUAA .................(((((.(.(((((.((((((((((((((..((((((......)))))).(-(....))....))--))))))))))))))))).).)))))..... ( -50.00, z-score = -3.49, R) >consensus GAUCUAAGUCCAGACCCUUC_CCUCCGAAAAGGGGGGCUUGGCACCUCCCCUUUUCUUGUCGAACACGCAGACAGACAACCC__CACACACAUACCCGAGUCAUUAACCAUUA ............(((.(.........(((((((((((........)))))))))))(((((.........)))))......................).)))........... (-16.00 = -16.24 + 0.24)


| Location | 17,828,000 – 17,828,102 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 69.87 |
| Shannon entropy | 0.49716 |
| G+C content | 0.50137 |
| Mean single sequence MFE | -34.22 |
| Consensus MFE | -15.30 |
| Energy contribution | -14.50 |
| Covariance contribution | -0.80 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.670407 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 17828000 102 + 22422827 GUCUGCGUGUUCGACAAGA--AAAGGGGAGGUGCCAAGCCC--CCCUUUUCGUAGG----GAAGGAUCUGGACUUAGAUCUAGG---UAACUAGAGCCAAGGCUUAUAAUUAC------- ((((..(.((((.....((--(((((((.(((.....))))--)))))))).(((.----.(.(((((((....)))))))...---)..)))))))).))))..........------- ( -35.00, z-score = -1.92, R) >droSec1.super_8 129814 102 + 3762037 GUCUGUGUGUCCGAAAAGA--AAAGGGGAGGUGCCAAGCCC--CCCUUUUCGGAGG----GAAGGGUCUGGACUUAGAUCUAGG---UUACUAGAGACAAGGCUUAUAAUUAC------- ((((.(((((((.....((--(((((((.(((.....))))--))))))))....(----((....))))))).....(((((.---...))))).)))))))..........------- ( -35.50, z-score = -1.76, R) >droYak2.chrX 16446690 102 + 21770863 GUCUGCGUGUUCGACAAGA--AAAGGGGAGGUGCCAAGCCC--CCCUCUUCGAAAC----GAAGGGUCUGGACUUAGAUCUAGG---UAACUAGCGCCAAGGCUUAUGAUUAC------- ((((((((.((((....((--(.(((((.(((.....))))--)))).)))....)----)))(((((((....)))))))...---......))))..))))..........------- ( -32.70, z-score = -1.04, R) >droEre2.scaffold_4690 8153425 102 + 18748788 GUCUGCGUGUUCGACAAGA--AAAGGGGAGGUGCCAAGCCC--CCCUUUUCAGAGC----GAAGGGUCUGGACUUAGAUCUAGG---UAACUAGAGCCAAGGCUUAUAAUUAU------- ((((.....((((....((--(((((((.(((.....))))--))))))))....)----)))((.((((((((((....))))---)..))))).)).))))..........------- ( -38.10, z-score = -2.83, R) >droVir3.scaffold_12928 4827852 120 - 7717345 UCACGCCCGUCUGGUGAGCUCACAGGAAAGCUCUCAAGCACAGCUCUGCUCUCAGGCCUUGACUAACCCAUACUUGCAUAGAGUACCCAACUCUAACUAAUCGUCAUGGCCCAUGCAUUC ....((..(..(((.(((((........))))))))..)...))..(((.....((((.((((.....((....))..((((((.....)))))).......)))).))))...)))... ( -29.80, z-score = -0.89, R) >consensus GUCUGCGUGUUCGACAAGA__AAAGGGGAGGUGCCAAGCCC__CCCUUUUCGGAGG____GAAGGGUCUGGACUUAGAUCUAGG___UAACUAGAGCCAAGGCUUAUAAUUAC_______ ((((....(((((((........((((..(((.....)))...))))((((.........)))).))).)))).....(((((.......)))))....))))................. (-15.30 = -14.50 + -0.80)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:54:32 2011