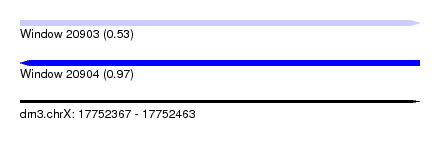
| Sequence ID | dm3.chrX |
|---|---|
| Location | 17,752,367 – 17,752,463 |
| Length | 96 |
| Max. P | 0.970259 |

| Location | 17,752,367 – 17,752,463 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 67.01 |
| Shannon entropy | 0.51399 |
| G+C content | 0.27763 |
| Mean single sequence MFE | -15.52 |
| Consensus MFE | -7.06 |
| Energy contribution | -7.62 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.532370 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
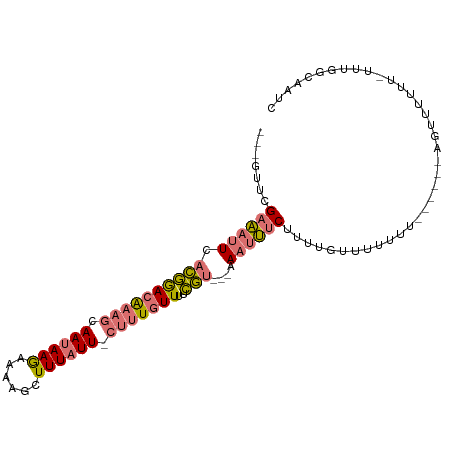
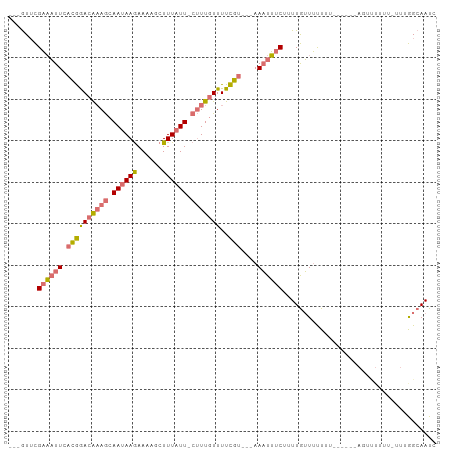
>dm3.chrX 17752367 96 + 22422827 ---GUUCGAAAUUCACGGACAUAGCAAUAAGAAAAGCUUUAUU-CUUUGUUUUCGU---AAAUUUCUUUUGUUUUUUUUAAUAAAGUUUUUUAUUUGGCAAUC ---(((((.......)))))...((.....(((((((((((((-.........((.---..........))........))))))))))))).....)).... ( -14.13, z-score = -0.90, R) >droSim1.chrX 13812364 91 + 17042790 ---GUUCGAAAUUCACGGACAAAGCAAUAAGAAAAGCUUUAUU-CUUUGUUUUCGU---AAAUUUCUUUUGUUUUUUU----AAAGUUUUUU-UUUGGCAAUC ---(((((.......)))))...((...(((((((((((((..-............---..................)----))))))))))-))..)).... ( -15.55, z-score = -1.26, R) >droSec1.super_8 62707 80 + 3762037 ---GUUCGAAAUUCACGGACAAAGCAAUAAGAAAAGCUUUAUU-CUUUGUUUUCGU---AAAUUUCUUUUGUUUUUUU----------------UUGGCAAUC ---....((((((.((((((((((.((((((......))))))-)))))))..)))---.))))))..(((((.....----------------..))))).. ( -16.50, z-score = -2.30, R) >droMoj3.scaffold_6328 3113471 97 - 4453435 UAGAUGUGUUUUUCUUAAACGAAAAAAAAAAAACACGUUUAUUUCCUUAUUUUUAUUCUAACUGCCGUUUAGUGCUGU------AAGCUGCGCCUAAGAAAUC (((((((((((((.((.........)).)))))))))))))..............((((...(((.(((((......)------)))).)))....))))... ( -15.90, z-score = -1.20, R) >consensus ___GUUCGAAAUUCACGGACAAAGCAAUAAGAAAAGCUUUAUU_CUUUGUUUUCGU___AAAUUUCUUUUGUUUUUUU______AGUUUUUU_UUUGGCAAUC .......((((((.((((((((((.((((((......)))))).)))))))..)))....))))))..................................... ( -7.06 = -7.62 + 0.56)

| Location | 17,752,367 – 17,752,463 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 67.01 |
| Shannon entropy | 0.51399 |
| G+C content | 0.27763 |
| Mean single sequence MFE | -16.52 |
| Consensus MFE | -6.71 |
| Energy contribution | -7.40 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.83 |
| SVM RNA-class probability | 0.970259 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
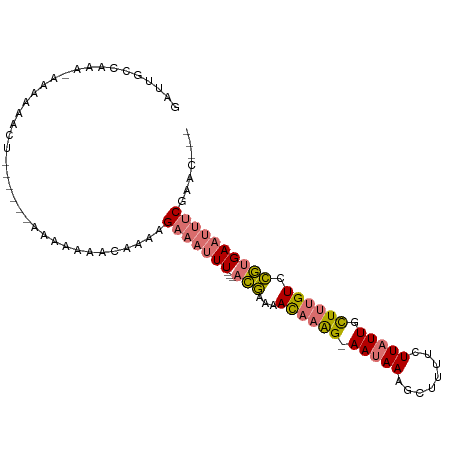
>dm3.chrX 17752367 96 - 22422827 GAUUGCCAAAUAAAAAACUUUAUUAAAAAAAACAAAAGAAAUUU---ACGAAAACAAAG-AAUAAAGCUUUUCUUAUUGCUAUGUCCGUGAAUUUCGAAC--- ........((((((....)))))).............(((((((---(((...(((.((-(((((........))))).)).))).))))))))))....--- ( -14.50, z-score = -2.42, R) >droSim1.chrX 13812364 91 - 17042790 GAUUGCCAAA-AAAAAACUUU----AAAAAAACAAAAGAAAUUU---ACGAAAACAAAG-AAUAAAGCUUUUCUUAUUGCUUUGUCCGUGAAUUUCGAAC--- ..........-..........----............(((((((---(((...((((((-(((((........))))).)))))).))))))))))....--- ( -17.50, z-score = -3.27, R) >droSec1.super_8 62707 80 - 3762037 GAUUGCCAA----------------AAAAAAACAAAAGAAAUUU---ACGAAAACAAAG-AAUAAAGCUUUUCUUAUUGCUUUGUCCGUGAAUUUCGAAC--- .........----------------............(((((((---(((...((((((-(((((........))))).)))))).))))))))))....--- ( -17.50, z-score = -3.60, R) >droMoj3.scaffold_6328 3113471 97 + 4453435 GAUUUCUUAGGCGCAGCUU------ACAGCACUAAACGGCAGUUAGAAUAAAAAUAAGGAAAUAAACGUGUUUUUUUUUUUUCGUUUAAGAAAAACACAUCUA ..(((((((.......((.------((.((........)).)).))........)))))))......(((((((((((.........)))))))))))..... ( -16.56, z-score = -0.92, R) >consensus GAUUGCCAAA_AAAAAACU______AAAAAAACAAAAGAAAUUU___ACGAAAACAAAG_AAUAAAGCUUUUCUUAUUGCUUUGUCCGUGAAUUUCGAAC___ .....................................(((((((...(((...((((((.(((((........))))).)))))).))))))))))....... ( -6.71 = -7.40 + 0.69)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:54:20 2011