| Sequence ID | dm3.chrX |
|---|---|
| Location | 17,700,310 – 17,700,424 |
| Length | 114 |
| Max. P | 0.527429 |

| Location | 17,700,310 – 17,700,424 |
|---|---|
| Length | 114 |
| Sequences | 9 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.29 |
| Shannon entropy | 0.09487 |
| G+C content | 0.36585 |
| Mean single sequence MFE | -24.23 |
| Consensus MFE | -21.34 |
| Energy contribution | -21.46 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.88 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.527429 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
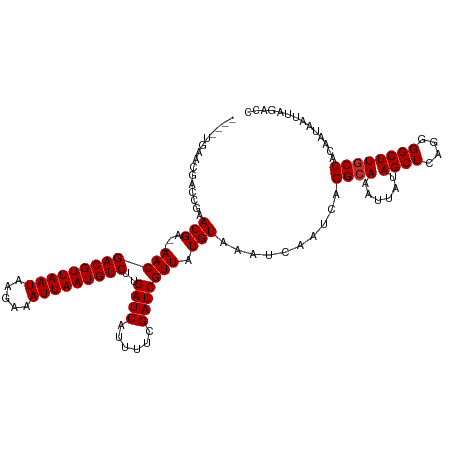
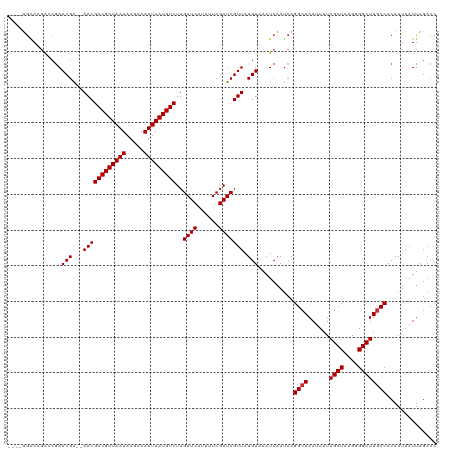
>dm3.chrX 17700310 114 + 22422827 ----UGAACGACCGAACGA--AACGACGUUAAUAAGAAAUUAAUGUCUUGAUCAUUUUCGAUCGUUAUGUAAAUCAAUCAGGCAAAUUAUAGCUCAGGGGCUUGCCACAAUAAUUAGACC ----(((...((..(((((--...(((((((((.....)))))))))((((......)))))))))..))...)))....((((......((((....)))))))).............. ( -24.60, z-score = -1.65, R) >droSim1.chrX_random 4534978 114 + 5698898 ----UGAACGACCGAACGA--AACGACGUUAAUAAGAAAUUAAUGUCUUGAUCAUUUUCGAUCGUUAUGUAAAUCAAUCAGGCAAAUUAUAGCUCAGGGGCUUGCCACAAUAAUUAGACC ----(((...((..(((((--...(((((((((.....)))))))))((((......)))))))))..))...)))....((((......((((....)))))))).............. ( -24.60, z-score = -1.65, R) >droSec1.super_8 10574 114 + 3762037 ----UGAACGACCGAACGA--AACGACGUUAAUAAGAAAUUAAUGUCUUGAUCAUUUUCGAUCGUUAUGUAAAUCAAUCAGGCAAAUUAUAGCUCAGGGGCUUGCCACAAUAAUUAGACC ----(((...((..(((((--...(((((((((.....)))))))))((((......)))))))))..))...)))....((((......((((....)))))))).............. ( -24.60, z-score = -1.65, R) >droYak2.chrX 16326073 114 + 21770863 ----UGAACAACCGAACGA--AACGACGUUAAUAAGAAAUUAAUGUCUUGAUCAUUUUCGAUCGUUAUGUAAAUCAAUCAGGCAAAUUAUAGCUCAGGGGCUUGCCACAAUAAUUAGACC ----...........(((.--((((((((((((.....)))))))))..((((......))))))).)))..........((((......((((....)))))))).............. ( -23.60, z-score = -1.67, R) >droEre2.scaffold_4690 8038851 114 + 18748788 ----UGAACGACCGAACGG--AACGACGUUAAUAAGAAAUUAAUGUCUUGAUCAUUUUCGAUCGUUAUGUAAAUCAAUCAGGCAAAUUAUAGCUCAGGGGCUUUCCACAAUAAUUAGACC ----..(((((.((((..(--(..(((((((((.....)))))))))....))...)))).)))))..............(((.((((((((((....)))).......)))))).).)) ( -21.81, z-score = -0.58, R) >droAna3.scaffold_13335 3000048 114 - 3335858 ----UAAAGGAGCGAACGA--AACGACGUUAAUAAGAAAUUAAUGUCUUGAUCAUUUUCGAUCGUUAUGUAAAUCAAUCAGGCAAAUUAUAGCUCAGGGGCUUGCCACAAUAAUUAGACC ----.....((..(((((.--((((((((((((.....)))))))))..((((......))))))).)))...))..)).((((......((((....)))))))).............. ( -24.30, z-score = -1.71, R) >dp4.chrXL_group1e 12090966 118 + 12523060 UGUAGGAAGGAACAAACGA--AACGACGUUAAUAAGAAAUUAAUGUCUUGAUCAUUUUCGAUCGUUAUGUAAAUCAAUCAGGCAAAUUAUAGCUCAGGGGCUUGCCACAAUAAUUAGACC (((..((..((((((((((--...(((((((((.....)))))))))((((......))))))))).)))...))..)).((((......((((....)))))))))))........... ( -25.10, z-score = -1.74, R) >droPer1.super_14 649016 118 - 2168203 UGUAGGAAGGAACGAACGA--AACGACGUUAAUAAGAAAUUAAUGUCUUGAUCAUUUUCGAUCGUUAUGUAAAUCAAUCAGGCAAAUUAUAGCUCAGGGGCUUGCCACAAUAAUUAGACC (((..((..((....(((.--((((((((((((.....)))))))))..((((......))))))).)))...))..)).((((......((((....)))))))))))........... ( -25.00, z-score = -1.39, R) >droWil1.scaffold_181096 4061633 116 + 12416693 ----UGGACAGAGGAACGACGAACGACGUUAAUAAGAAAUUAAUGUCUUGAUCAUUUUCGAUCGUUAUGUAAAUCAAUCAGGCAAAUUAUAGCUCAGGGGCUUGCCACAAUAAUUAGACC ----.((...((.((...(((((((((((((((.....)))))))))..((((......))))))).)))...))..)).((((......((((....))))))))............)) ( -24.50, z-score = -1.24, R) >consensus ____UGAACGACCGAACGA__AACGACGUUAAUAAGAAAUUAAUGUCUUGAUCAUUUUCGAUCGUUAUGUAAAUCAAUCAGGCAAAUUAUAGCUCAGGGGCUUGCCACAAUAAUUAGACC ...............(((...((((((((((((.....)))))))))..((((......))))))).)))..........((((......((((....)))))))).............. (-21.34 = -21.46 + 0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:54:14 2011