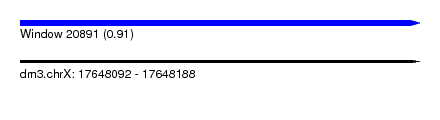
| Sequence ID | dm3.chrX |
|---|---|
| Location | 17,648,092 – 17,648,188 |
| Length | 96 |
| Max. P | 0.906399 |

| Location | 17,648,092 – 17,648,188 |
|---|---|
| Length | 96 |
| Sequences | 11 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 88.82 |
| Shannon entropy | 0.22594 |
| G+C content | 0.34422 |
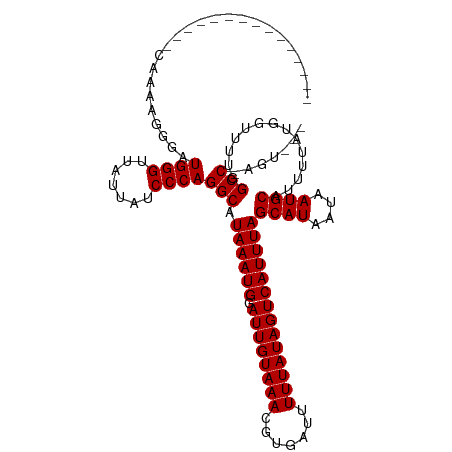
| Mean single sequence MFE | -26.05 |
| Consensus MFE | -19.80 |
| Energy contribution | -19.80 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.76 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.906399 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chrX 17648092 96 + 22422827 --------------CGAAUGGG-AUGGGUUAUUAUCCCAGGCAUAAAUGGAUUGUAAACGUGAUUUUUAUAGUCAUUUAGCAUAAUAAUGCAUUUUAUGGUUUUGGCC-AGU--- --------------....((((-((((....))))))))(((.((((((.((((((((.......))))))))))))))((((....))))..............)))-...--- ( -26.00, z-score = -2.46, R) >droSec1.super_17 1494386 96 + 1527944 ---------------CGAAUGGGAUGGGUUAUUAUCCCAGGCAUAAAUGGAUUGUAAACGUGAUUUUUAUAGUCAUUUAGCAUAAUAAUGCAUUUUAUGGUUUUGGCC-AGU--- ---------------....((((((((....))))))))(((.((((((.((((((((.......))))))))))))))((((....))))..............)))-...--- ( -26.00, z-score = -2.46, R) >droYak2.chrX 16276184 96 + 21770863 ---------------CGAAUGGGAUGGGUUAUUAUCCCAGGCAUAAAUGGAUUGUAAACGUGAUUUUUAUAGUCAUUUAGCAUAAUAAUGCAUUUUAUGGUUUUGGCC-AGU--- ---------------....((((((((....))))))))(((.((((((.((((((((.......))))))))))))))((((....))))..............)))-...--- ( -26.00, z-score = -2.46, R) >droEre2.scaffold_4690 7983222 96 + 18748788 ---------------CGAAUGGGAUGGGUUAUUAUCCCAGGCAUAAAUGGAUUGUAAACGUGAUUUUUAUAGUCAUUUAGCAUAAUAAUGCAUUUUAUGGUUUUGGCC-AGU--- ---------------....((((((((....))))))))(((.((((((.((((((((.......))))))))))))))((((....))))..............)))-...--- ( -26.00, z-score = -2.46, R) >droAna3.scaffold_13335 2945614 112 - 3335858 ACGGAGUCCGGACUCCGAAUGGGAUGGGUUAUUAUCCCAGGCAUAAAUGGAUUGUAAACGUGAUUUUUAUAGUCAUUUAGCAUAAUAAUGCAUUUUAUGGUUUUGGCC-AGUC-- .((((((....))))))..((((((((....))))))))(((.((((((.((((((((.......))))))))))))))((((....))))..............)))-....-- ( -35.60, z-score = -3.17, R) >dp4.chrXL_group1e 12036110 108 + 12523060 ----UGGGGAAUGGGGAAUGGGCAUGGGUUAUUAUCCCAGGCAUAAAUGGAUUGUAAACGUGAUUUUUAUAGUCAUUUAGCAUAAUAAUGCAUUUUAUGGUUUUAGCCCAGU--- ----..............(((((.((((.......))))..((((((..(((((((((.......))))))))).....((((....))))..))))))......)))))..--- ( -27.70, z-score = -1.18, R) >droPer1.super_14 593800 101 - 2168203 -----------UGGGGAAUGGGCAUGGGUUAUUAUCCCAGGCAUAAAUGGAUUGUAAACGUGAUUUUUAUAGUCAUUUAGCAUAAUAAUGCAUUUUAUGGUUUUAGCCCAGU--- -----------.......(((((.((((.......))))..((((((..(((((((((.......))))))))).....((((....))))..))))))......)))))..--- ( -27.70, z-score = -1.93, R) >droWil1.scaffold_181096 3999828 98 + 12416693 ---------------CAAAAGAGAUGGGUUAUUAUCCCAGGCAUAAAUGGAUUGUAAACGUGAUUUUUAUAGUCAUUUAGCAUAAUAAUGCAUUUUAUGGUUUUAGCC-AGAGU- ---------------.....((((((.((((((((.....((.((((((.((((((((.......)))))))))))))))))))))))).)))))).((((....)))-)....- ( -23.30, z-score = -2.54, R) >droVir3.scaffold_12928 5062095 95 - 7717345 --------------UCAAAAGAGAUGGGUUAUUAUCCCAGGCAUAAAUGGAUUGUAAACGUGAUUUUUAUAGUCAUUUAGCAUAAUAAUGCAUUUUAUGG-UUU-GCC-AGU--- --------------......((((((.((((((((.....((.((((((.((((((((.......)))))))))))))))))))))))).)))))).(((-...-.))-)..--- ( -22.40, z-score = -2.05, R) >droMoj3.scaffold_6473 2514968 96 - 16943266 --------------UCAAAAGAGAUGGGUUAUUAUCCCAGGCAUAAAUGGAUUGUAAACGUGAUUUUUAUAGUCAUUUAGCAUAAUAAUGCAUUUUAUGGCUUU-GCC-AGU--- --------------......((((((.((((((((.....((.((((((.((((((((.......)))))))))))))))))))))))).)))))).((((...-)))-)..--- ( -23.50, z-score = -2.00, R) >droGri2.scaffold_14853 6736953 98 + 10151454 --------------UCAAAAGAGAUGGGUUAUUAUCCCAGGCAUAAAUGGAUUGUAAACGUGAUUUUUAUAGUCAUUUAGCAUAAUAAUGCAUUUUAUGGUCU--GCC-AGAGUC --------------......((((((.((((((((.....((.((((((.((((((((.......)))))))))))))))))))))))).)))))).(((...--.))-)..... ( -22.40, z-score = -1.68, R) >consensus _______________CAAAAGGGAUGGGUUAUUAUCCCAGGCAUAAAUGGAUUGUAAACGUGAUUUUUAUAGUCAUUUAGCAUAAUAAUGCAUUUUAUGGUUUUGGCC_AGU___ ........................((((.......))))(((.((((((.((((((((.......))))))))))))))((((....))))..............)))....... (-19.80 = -19.80 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:54:09 2011