| Sequence ID | dm3.chrX |
|---|---|
| Location | 17,643,976 – 17,644,069 |
| Length | 93 |
| Max. P | 0.676557 |

| Location | 17,643,976 – 17,644,069 |
|---|---|
| Length | 93 |
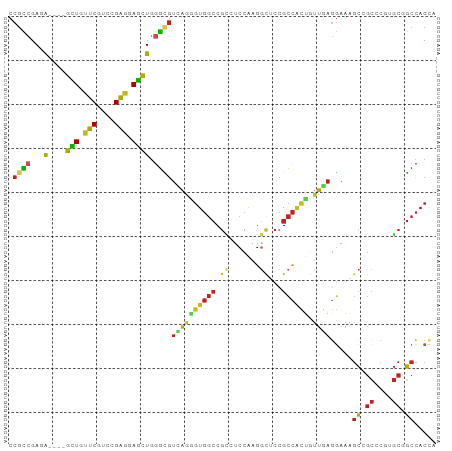
| Sequences | 11 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 75.87 |
| Shannon entropy | 0.52594 |
| G+C content | 0.65840 |
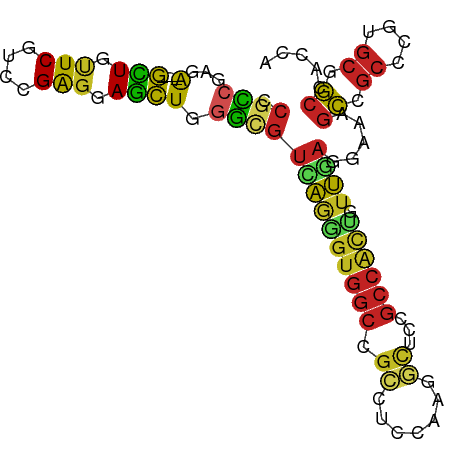
| Mean single sequence MFE | -41.56 |
| Consensus MFE | -18.28 |
| Energy contribution | -17.53 |
| Covariance contribution | -0.75 |
| Combinations/Pair | 1.74 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.676557 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 17643976 93 - 22422827 CCGCCGAGA----GCUGUUCGUCCGAGGAGCUGGGCGUCAAAGUGGCCGCCUCCAAGGCUCCGCCACUGUUGAGGAAAGCCGCCCUCGCGGCCACCA ..((((.((----(((.(((....))).))).((((.((((((((((.(((.....)))...)))))).))))((....)))))))).))))..... ( -40.90, z-score = -1.20, R) >droSim1.chrX 13740142 93 - 17042790 CCGCCGACA----GCUGUUCGUCCGAGGAGCUGGGCGUCAAAGUGGCCGCCUCCAAGGCUCCGCCACUGUUGAGGAAAGCCGCCCUCGCGGCCACCA .((((..((----(((.(((....))).)))))))))((((((((((.(((.....)))...)))))).))))((...(((((....)))))..)). ( -43.60, z-score = -2.15, R) >droSec1.super_17 1490359 93 - 1527944 CCGCCGACA----GCUGUUCGUCCGAGGAGCUGGGCGUUAAAGUGGCCGCCUCCAAGGCUCCGCCACUGUUGAGGAAAGCCGCCCUCGCGGCCACCA .((((..((----(((.(((....))).))))))))).....((((((((......(((...)))......((((........)))))))))))).. ( -42.60, z-score = -2.04, R) >droYak2.chrX 16271945 93 - 21770863 CCGCCGAGA----GCUGUUCGUCCGAGGAGCUGGGCGUCAGAGUGGCCGCCUCCAAGGCUCCGCCACUAUUGAGGAAAGCCGCCCUGGCGGCCACCA .((((...(----(((.(((....))).)))).))))...(.(((((((((.....((((...((.(....).))..)))).....)))))))))). ( -44.30, z-score = -1.88, R) >droEre2.scaffold_4690 7979317 93 - 18748788 CCCCCGAGA----GCUGUUCGUCCGAGGAGCUGGGCGUCAAUGUGGCCGCCUCCAAGGCUCCGCCACUGUUGAGGAAAGCCGCCCUGGCGGCCACCA ..(((...(----(((.(((....))).))))))).......(((((((((.....((((...((.(....).))..)))).....))))))))).. ( -40.70, z-score = -1.16, R) >dp4.chrXL_group1e 413738 93 + 12523060 CCGCCGACA----GCUGCUCGUCUGAGGAGCUGGGCGUCAGGGUGGCCGGGUCCAAGGCCCCGCCGCUGUUGAGGAAGGCGGCACGGGCGGCCACCA .((((..((----(((.(((....))).)))))))))....((((((((....)...((((.((((((.........))))))..))))))))))). ( -52.40, z-score = -1.59, R) >droWil1.scaffold_181096 8331972 93 + 12416693 CCGCCGAUA----GUUGUUCAUCCGAUGAGCUGGGUGUCAGGGUGGCCGUAUCCAAGUUGCCGCCAUUAUUGAGGAAUGCAGCACGAGCCGCCACAA .....((((----.((((((((...)))))).)).))))..(((((((((......(((((..((........))...)))))))).)))))).... ( -29.20, z-score = -0.15, R) >droVir3.scaffold_12928 2567474 93 - 7717345 CCGCCGAUA----GCUGCUCGUCCGAGGAGCUCGGUGUCAGGGUGGCCGCAUCCAAGGUGCCGCCGCUGUUCAGAAAGGCGGCGCGUGCCGCCACAA .((((((..----(((.(((....))).))))))))).....(((((.((((.....((((((((.((....))...)))))))))))).))))).. ( -48.60, z-score = -2.65, R) >droMoj3.scaffold_6473 14333549 93 - 16943266 CCGUGGACA----GCUGCUCGUCCGAGGAGCUGGGCGUCAAGGUGGCCGCAUCCAAGGUGCCGCCGCUGUUCAAGAAGGCGGCCCGUGCCGCCACCA .(((...((----(((.(((....))).))))).)))....((((((.((((....((.((((((.((.....))..)))))))))))).)))))). ( -48.50, z-score = -2.32, R) >droGri2.scaffold_15203 9689981 93 - 11997470 CCGCCGAGA----GCUGCUCAUCCGAGGAGCUGGGCGUUAGUGUGGCCGCAUCCAAGGUGCCGCCAUUGUUAAGGAAGGCGGCGCGUGCAGCCACCA .((((...(----(((.(((....))).)))).))))...(.(((((.((((.....((((((((.((......)).)))))))))))).)))))). ( -44.80, z-score = -2.21, R) >apiMel3.Group3 10274558 94 + 12341916 CCGACGAGGCAACAUUGCUCAAUUGGGAAAUUGAUAUUUGGGAU---CCUUUCUGAGAUUUUGAUUCCGACACUUCGUCGUCAGCGUACCUCAACAG .((((((((.........((((((....))))))...(((((((---(...((...))....))))))))..))))))))................. ( -21.60, z-score = -0.24, R) >consensus CCGCCGAGA____GCUGUUCGUCCGAGGAGCUGGGCGUCAGGGUGGCCGCCUCCAAGGCUCCGCCACUGUUGAGGAAAGCCGCCCGUGCGGCCACCA .............(((.(((....))).)))..(((.((((((((((.((.......))...)))))).))))........((....)).))).... (-18.28 = -17.53 + -0.75)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:54:08 2011