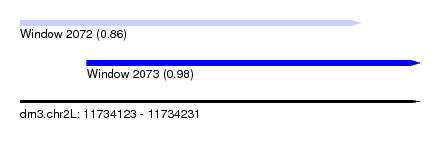
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 11,734,123 – 11,734,231 |
| Length | 108 |
| Max. P | 0.983202 |

| Location | 11,734,123 – 11,734,215 |
|---|---|
| Length | 92 |
| Sequences | 9 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 60.45 |
| Shannon entropy | 0.79194 |
| G+C content | 0.50793 |
| Mean single sequence MFE | -25.08 |
| Consensus MFE | -10.58 |
| Energy contribution | -11.77 |
| Covariance contribution | 1.19 |
| Combinations/Pair | 1.42 |
| Mean z-score | -0.91 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.859471 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 11734123 92 + 23011544 -------UCCGAAAAUAAAACGCCAAUCUCU--UAUAGGUACAU-AGACUAACCUUUUGCCUCUA-----AGGCGACCUUGUGGC-GAAACUUUUCACAUGGCGUCAA- -------............((((((......--....(((....-......)))(((((((.(.(-----(((...))))).)))-)))).........))))))...- ( -20.70, z-score = -1.81, R) >droSim1.chr2L 11536789 93 + 22036055 -------UCCGAAAAUAAAAUGCCAACCCCUA-CAUAGGUACAU-AGACUAACCUUUUGCCUCUA-----GGGCGACCUUGUGGC-GAAACUUUUCACAGGGCGUCAA- -------............(((((.(((....-....)))....-.........(((((((.(.(-----((....))).).)))-))))..........)))))...- ( -22.10, z-score = -1.01, R) >droSec1.super_3 7124760 93 + 7220098 -------UCCGAAAAUAAAAUGCCAACCCCUA-CAUAGGUACUU-AGACUAACCUUUUGCCUCUG-----GGGCGACCUUGUGGC-GAAACUUUUCACAGGGCGUCAA- -------............(((((.(((....-....)))....-.........(((((((.(.(-----((....))).).)))-))))..........)))))...- ( -22.10, z-score = -0.46, R) >droYak2.chr2L 8152275 90 + 22324452 --------UCAGAAGAAAAAUGCCAACCCCU---AUAAGUACAU-GGACCAACCUUUUGCCUCUA-----GGGCGACCUUGUGGC-GAAACUUUGCACAGGGCGCCAA- --------.((((((.......(((((....---....))...)-))......))))))......-----.((((.(((((((.(-((....))))))))))))))..- ( -23.92, z-score = -0.87, R) >droEre2.scaffold_4929 12958544 70 - 26641161 --------UCAGAAAAAAAAUGCCAACCCC----AUAGGUACAU-AGACUAACCUUUUGCCUCU------GGGCGACCUUGUGGC-GAAA------------------- --------............(((((.((((----(.(((((...-((......))..))))).)------))).)......))))-)...------------------- ( -17.10, z-score = -1.59, R) >droAna3.scaffold_12916 9442485 99 - 16180835 -------CCACCACGCACACAGCCCACCACAAAAAUAGGUACGCCAGGCCAACCUUUGGGCUCUAGGUC-GGGCGACCUUGCGGC-AAAACUUUCCACAGGGCGUCCU- -------.......((....((((((...........((....))(((....))).))))))....)).-(((((.(((((.((.-((....)))).)))))))))).- ( -29.70, z-score = -0.42, R) >droPer1.super_1 4074954 107 + 10282868 UGCCCCCCUUCCACCCCAUUUCACACCCCUCUUCGCACUGAAGGUGGGAUAACCUUUUGGCUCUAGGUC-GAGCGACCUUGUGGC-AAAACUUUCCGCAGGGCGCAAAA .........((.(((..........(((.(((((.....))))).)))....((....)).....))).-))(((.((((((((.-((....))))))))))))).... ( -30.60, z-score = -0.49, R) >dp4.chr4_group3 2600736 107 + 11692001 UGCCCCCCUUCCACCCCCUUUCACCCCCCUGUUCGCACUGAAGGUGGGCCAACCUUUUGGCUCUAGGUC-GAGCGACCUUGUGGC-AAAACUUUCCGCAGGGCGCAAAA .((((.(((((..........((......))........))))).)))).......(((((.....)))-))(((.((((((((.-((....))))))))))))).... ( -32.67, z-score = -0.35, R) >droWil1.scaffold_180772 5204068 92 - 8906247 ----------------AGGCAGCUAACCUUUUGGCCCUCUUCUCUGGUCCCUCUCAAUCUGUCUAGGUUUUUGCGACCUUGUGGUGAAAACUUUCCGCAGGGUGCAAA- ----------------((((((..........((.((........)).))........)))))).....(((((.(((((((((.((....)).))))))))))))))- ( -26.87, z-score = -1.18, R) >consensus ________CCCAAAACAAAAUGCCAACCCCU__CAUAGGUACAU_AGACUAACCUUUUGCCUCUA_____GGGCGACCUUGUGGC_GAAACUUUCCACAGGGCGCCAA_ .......................................................................((((.((((((((..........))))))))))))... (-10.58 = -11.77 + 1.19)

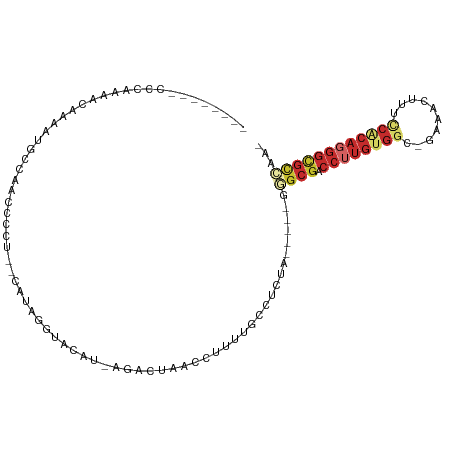
| Location | 11,734,141 – 11,734,231 |
|---|---|
| Length | 90 |
| Sequences | 12 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 59.19 |
| Shannon entropy | 0.84084 |
| G+C content | 0.48608 |
| Mean single sequence MFE | -25.03 |
| Consensus MFE | -11.31 |
| Energy contribution | -11.07 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.73 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.13 |
| SVM RNA-class probability | 0.983202 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr2L 11734141 90 + 23011544 ----AUCUCU---UAUAGGUACAU-AGACUAAC-CUUUUGCCUCUA----A-----GGCGACCUUGUGG-CGAAACUUUUCACAUGGCGUCAACUUUUUUCCCAUUUUA---- ----.....(---((.(((((...-((......-))..))))).))----)-----((((.((.(((((-.........))))).))))))..................---- ( -16.80, z-score = -1.20, R) >droSim1.chr2L 11536807 91 + 22036055 ----ACCCCUA--CAUAGGUACAU-AGACUAAC-CUUUUGCCUCUA----G-----GGCGACCUUGUGG-CGAAACUUUUCACAGGGCGUCAACUUUUUUCCCAUUUCA---- ----..(((((--...(((((...-((......-))..))))).))----)-----))((.((((((((-.........))))))))))....................---- ( -21.20, z-score = -1.39, R) >droSec1.super_3 7124778 91 + 7220098 ----ACCCCUA--CAUAGGUACUU-AGACUAAC-CUUUUGCCUCUG----G-----GGCGACCUUGUGG-CGAAACUUUUCACAGGGCGUCAACUUUUUUCCCAUUUCA---- ----.((((..--...(((((...-((......-))..)))))..)----)-----))((.((((((((-.........))))))))))....................---- ( -21.70, z-score = -0.99, R) >droYak2.chr2L 8152292 89 + 22324452 ----ACCCCU----AUAAGUACAU-GGACCAAC-CUUUUGCCUCUA----G-----GGCGACCUUGUGG-CGAAACUUUGCACAGGGCGCCAACUUUUUUCCCAUUUAA---- ----....((----(...(((...-((.....)-)...)))...))----)-----((((.(((((((.-(((....))))))))))))))..................---- ( -22.40, z-score = -1.13, R) >droEre2.scaffold_4929 12958561 69 - 26641161 ----ACCCC-----AUAGGUACAU-AGACUAAC-CUUUUGCCUCU-----G-----GGCGACCUUGUGG-CGAAACUUUUCUUCCCUUUUA---------------------- ----..(((-----(.(((((...-((......-))..))))).)-----)-----))((.((....))-))...................---------------------- ( -14.80, z-score = -1.35, R) >droAna3.scaffold_12916 9442503 93 - 16180835 ----ACCACAAA-AAUAGGUACGCCAGGCCAAC-CUUUGGGCUCUAGGUCG-----GGCGACCUUGCGG-CAAAACUUUCCACAGGGCGUCCUGUUUUUUUCUUC-------- ----.....(((-((((((.(((((..(((..(-(...))((...(((((.-----...))))).))))-)....((......))))))))))))))))......-------- ( -31.60, z-score = -2.09, R) >droPer1.super_1 4074974 105 + 10282868 UCACACCCCUCUUCGCACUGAAGGUGGGAUAAC-CUUUUGGCUCUAGGUCG-----AGCGACCUUGUGG-CAAAACUUUCCGCAGGGCGCAAAAGUUUUUUUUCC-CGUCGUC .....(((.(((((.....))))).))).....-((((((((((((((((.-----...))))).((((-.((....))))))))))).))))))..........-....... ( -31.10, z-score = -0.69, R) >dp4.chr4_group3 2600756 106 + 11692001 UCACCCCCCUGUUCGCACUGAAGGUGGGCCAAC-CUUUUGGCUCUAGGUCG-----AGCGACCUUGUGG-CAAAACUUUCCGCAGGGCGCAAAAGUUUUUUUUCCUCAUCGUC (((((..(((...........))).(((((((.-...)))))))..))).)-----)(((.((((((((-.((....)))))))))))))....................... ( -29.90, z-score = -0.05, R) >droWil1.scaffold_180772 5204077 94 - 8906247 ----ACCUU------UUGGCCCUCUUCUCUGGUCCCUCUCAAUCUGUCUAGGUUUUUGCGACCUUGUGGUGAAAACUUUCCGCAGGGUGCAAAGUUUCUCCCUU--------- ----.....------..((.((........)).))...............((.((((((.(((((((((.((....)).))))))))))))))).....))...--------- ( -23.20, z-score = -1.19, R) >droVir3.scaffold_12963 6309102 97 + 20206255 ----GCCCCGCACCGCAGUC----UAACCUAAC-CUUUUGGCCAUGUUAGGUUUU-AGCGACCUUGCAGUCAAAACUUUCUGCAGGGCG-GAAAGUUUUUAGCAUUUU----- ----...((((..(((....----.((((((((-...........))))))))..-.)))..(((((((..........))))))))))-).................----- ( -27.60, z-score = -1.40, R) >droMoj3.scaffold_6500 25455221 98 + 32352404 ----GCCCCGU--CGCCGUC----UAACCUAAC-CCUUUGGCCAUACUAGGUUUUUGGCGACCUUGCGGCCAAAACUUUCUGCAGGGCGCAAAAGUUUUUCGCAUUUUA---- ----.....((--(((((..----.((((((..-.............))))))..)))))))..(((((..((((((((.(((.....)))))))))))))))).....---- ( -30.46, z-score = -1.82, R) >droGri2.scaffold_15252 9555821 98 + 17193109 ----GCCCCAC--CGCAGUCG---CAGUCUAUCGUUUUUGGCCAUUCUAGGUUU--AGCGACCUUGCGGCCAAAACUUUCUGCAGGGCGAAAAGUUUUUGUGGAUUUUU---- ----......(--((((((((---(((....(((((...((((......)))).--)))))..))))))).((((((((.(((...))).)))))))))))))......---- ( -29.60, z-score = -1.28, R) >consensus ____ACCCCU___CAUAGGUACAU_AGACUAAC_CUUUUGGCUCUA____G_____GGCGACCUUGUGG_CAAAACUUUCCGCAGGGCGCAAAAUUUUUUCCCAUUUUA____ .........................................................(((.((((((((..........)))))))))))....................... (-11.31 = -11.07 + -0.24)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:33:28 2011