| Sequence ID | dm3.chrX |
|---|---|
| Location | 17,633,934 – 17,634,041 |
| Length | 107 |
| Max. P | 0.529529 |

| Location | 17,633,934 – 17,634,041 |
|---|---|
| Length | 107 |
| Sequences | 8 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.79 |
| Shannon entropy | 0.44879 |
| G+C content | 0.58014 |
| Mean single sequence MFE | -42.38 |
| Consensus MFE | -24.08 |
| Energy contribution | -23.12 |
| Covariance contribution | -0.95 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.529529 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 17633934 107 - 22422827 -------------GGAUAGUGCCUGCUGUUUGGGCGGCGGGGGCGUGGCCAGGACAAUGUCGUUCUCCUCAUUGAAAUGAGUCCUGGGCGCAGCCGCAUUUGGUAUGUUCUGGCCGUUUG -------------.....((.((((((((....)))))))).))..((((((((((.((.(((((..(((((....)))))....)))))))((((....)))).))))))))))..... ( -47.20, z-score = -2.39, R) >droGri2.scaffold_15203 9677615 105 - 11997470 GGGAUCGUGAGCGAGAGAGCGCCUGCUGUUUAGGCGGC------GUUGCCAUCACAAUGUCGUUAUCUUCAUUGAAAUGAGUGCGUGGCGCA----CACCUGGCGCCAUCUGGCG----- ..........((.(((..(((((.(((((....)))))------(((((((((((....(((((.((......)))))))))).)))))).)----)....)))))..))).)).----- ( -38.70, z-score = -0.33, R) >droMoj3.scaffold_6473 14320954 105 - 16943266 GUGCGGGUGAGAGCGAGAGCGCCUGCUGCUUGGGCGGC------GUUGCCAUCACAAUGUCGUUAUCUUCAUUGAAAUGAGUGCGUGGCGCA----CACCUGGCGCCAUCUGGCG----- ...((((((...((....))((((((.((((.((((((------((((......)))))))))).((......))...))))))).)))...----)))))).((((....))))----- ( -41.20, z-score = -0.04, R) >droVir3.scaffold_12928 2554309 105 - 7717345 GUGCGGGUGAGAGCGAGAGCGCCUGCUGUUUGGGCGGC------GUUGCCAUCACAAUGUCGUUAUCUUCAUUGAAGUGUGUGCGUGGCGCA----CACCUGGCGCCAUCUGGCG----- ...((((((...((....))((((.......)))).((------((..(((.((((...(((..........)))..)))))).)..)))).----)))))).((((....))))----- ( -41.40, z-score = 0.02, R) >droEre2.scaffold_4690 7969258 107 - 18748788 -------------GGAUAAUGCCUGCUGUUUUGGCGGCGGGGGCGUGGCCAGGACAAUGUCGUUCUCCUCAUUGAAAUGAGUCCUGGGCGCAGCCGUAUUUGGUAUGUUCUGGGUGUUUG -------------........((((((((....))))))))((((((.(((((((....((((((........).)))))))))))).))).))).((((..(......)..)))).... ( -40.90, z-score = -1.76, R) >droYak2.chrX 16261736 107 - 21770863 -------------GGAUAAUGCCUGCUGUUUGGGCGGCGGGGGCGUGGCCAGGACAAUGUCGUUCUCCUCAUUGAAAUGAGUCCUGGGCGCAGCCGUAUUUGGUAUAUUCUGGCUGUUUG -------------(((((...((((((((....))))))))((((((.(((((((....((((((........).)))))))))))).))).)))..........))))).......... ( -40.50, z-score = -1.48, R) >droSec1.super_17 1480409 107 - 1527944 -------------GGAUAGAGCCUGCUGUUUGGGCGGCGGGGGAGUGGCCAGGACAAUGUCGUUCUCCUCAUUGAAAUGAGUCCUGGGCGCAGCCGCAUUUGGUAUGUUCUGGCUGUUUG -------------........((((((((....)))))))).(((..(((((((((.((.(((((..(((((....)))))....)))))))((((....)))).)))))))))..))). ( -43.90, z-score = -1.98, R) >droSim1.chr3L 15823618 107 + 22553184 -------------GGAUAGUGCCUGCUGUUUGGGCGGCGGGGGCGUGGCCAGGACAAUGUCGUUCUCCUCAUUGAAAUGUGUCCUGGGCGCAGCCGCAUUUGGUAUGUUCUGGCUGUUUG -------------.....((.((((((((....)))))))).))(((.((((((((......(((........)))...)))))))).)))((((((((.....))))...))))..... ( -45.20, z-score = -2.17, R) >consensus _____________GGAUAGUGCCUGCUGUUUGGGCGGCGGGGGCGUGGCCAGGACAAUGUCGUUCUCCUCAUUGAAAUGAGUCCUGGGCGCAGCCGCAUUUGGUAUGUUCUGGCUGUUUG .............(((..(((((...(((...(((.(((....))).)))...))).((.(((((..(((((....)))))....))))))).........)))))..)))......... (-24.08 = -23.12 + -0.95)

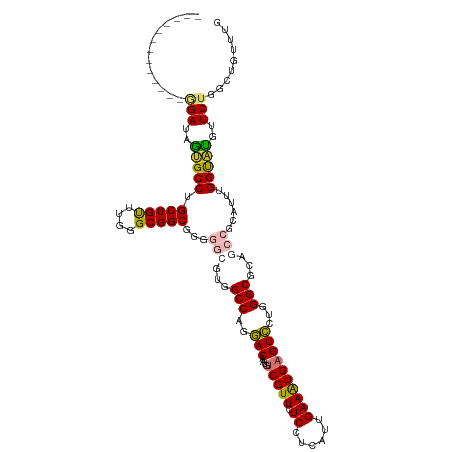

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:54:05 2011