
| Sequence ID | dm3.chrX |
|---|---|
| Location | 17,627,706 – 17,627,806 |
| Length | 100 |
| Max. P | 0.841727 |

| Location | 17,627,706 – 17,627,806 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 73.77 |
| Shannon entropy | 0.45509 |
| G+C content | 0.36750 |
| Mean single sequence MFE | -21.18 |
| Consensus MFE | -13.72 |
| Energy contribution | -15.76 |
| Covariance contribution | 2.04 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.841727 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 17627706 100 + 22422827 CUGCAUUUAUCGAUUGGCGCCU-------ACAUUUGAGUGGAAAACUCUGGCUUUAAAGUCGAGCUAUCUGAUAGAUAAAUUUAGAUGUAACACUAUAUUGAAAUAA- ....((((((((..((((....-------......((((.....))))(((((....))))).))))..))))))))..((((.((((((....)))))).))))..- ( -21.00, z-score = -1.39, R) >droSim1.chrX 13731309 94 + 17042790 CUGCAUUUAUCGAUUGGCGCCU-------ACAUUUAAGAGGAAAACUCUGGCUUUAAAGUCGAGCUAUCUGCCAGAUAAAUGGAGAUUUUCAACUAUAAUA------- ((.((((((((...((((....-------.......((((.....))))(((((.......)))))....)))))))))))).))................------- ( -23.60, z-score = -2.05, R) >droSec1.super_17 1473142 93 + 1527944 CUGCAUUUAUCGAUUGGCGCCU-------ACAUUUAAGAGGAAAACUCUGGCUU-AAAGUCGAGCUAUCUGCCAGAUAAAUAAGGAUUUUCAACUAUAAUA------- ((..(((((((...((((....-------.......((((.....))))(((((-......)))))....)))))))))))..))................------- ( -19.50, z-score = -1.01, R) >droYak2.chrX 16255463 107 + 21770863 CUGCAUUUAUCGAUUGGCGCCUUAAUUAAGAGGAAAAGAGGAAAACUCUGGCUGCAAAGUCGAGCUAUCUGCCAGAUAAAUGU-CAUUUGUAACUAUAUUAUUCAUAU ..(((((((((...((((.((((......))))......(((...(((.((((....)))))))...))))))))))))))))-........................ ( -24.70, z-score = -1.19, R) >droEre2.scaffold_4690 7963053 81 + 18748788 CUGCAUUUAUCGAUUGGCGCCCCA-----GACUUAAGAGGGAAAACUC-------------GUGCUAUCUGCCAGAUAAAUGU-GAUUUUAAACUAUAUU-------- ..(((((((((...(((((....(-----(.(....(((......)))-------------..)))...))))))))))))))-................-------- ( -17.10, z-score = -1.14, R) >consensus CUGCAUUUAUCGAUUGGCGCCU_______ACAUUUAAGAGGAAAACUCUGGCUU_AAAGUCGAGCUAUCUGCCAGAUAAAUGU_GAUUUUAAACUAUAUUA_______ (((((((((((...((((..................((((.....))))((((....)))).........)))))))))))))))....................... (-13.72 = -15.76 + 2.04)

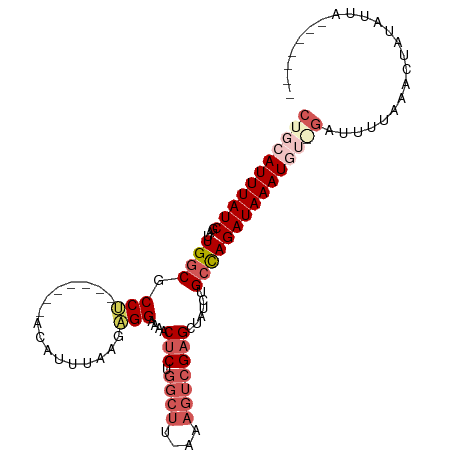
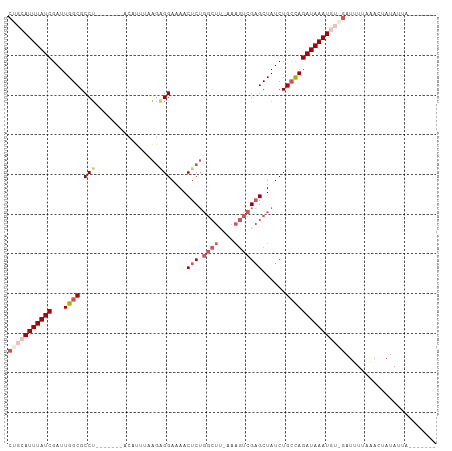
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:54:03 2011