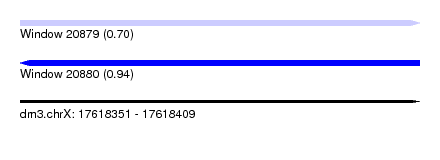
| Sequence ID | dm3.chrX |
|---|---|
| Location | 17,618,351 – 17,618,409 |
| Length | 58 |
| Max. P | 0.943749 |

| Location | 17,618,351 – 17,618,409 |
|---|---|
| Length | 58 |
| Sequences | 7 |
| Columns | 62 |
| Reading direction | forward |
| Mean pairwise identity | 82.64 |
| Shannon entropy | 0.35942 |
| G+C content | 0.43407 |
| Mean single sequence MFE | -12.76 |
| Consensus MFE | -9.56 |
| Energy contribution | -9.56 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.702135 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
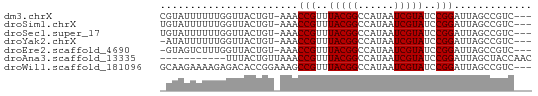
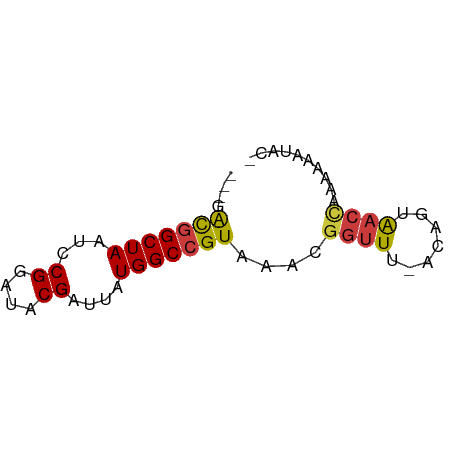
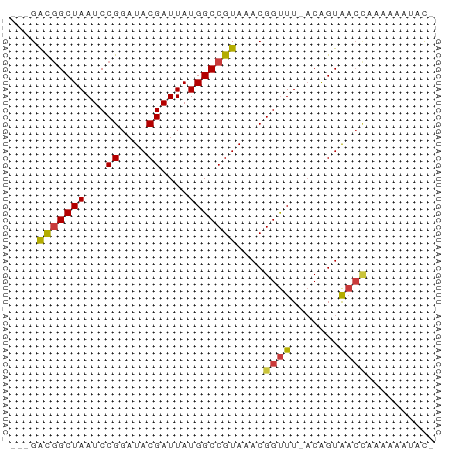
>dm3.chrX 17618351 58 + 22422827 CGUAUUUUUUGGUUACUGU-AAACCGUUUACGGCCAUAAUCGUAUCCGGAUUAGCCGUC--- ..........((((.....-.))))....(((((..(((((.......)))))))))).--- ( -12.40, z-score = -0.80, R) >droSim1.chrX 3680692 58 - 17042790 UGUAUUUUUUGGUUACUGU-AAACCGUUUACGGCCAUAAUCGUAUCCGGAUUAGCCGUC--- ..........((((.....-.))))....(((((..(((((.......)))))))))).--- ( -12.40, z-score = -0.92, R) >droSec1.super_17 1464064 58 + 1527944 UGUAUUUUUUGGUUACUGU-AAACCGUUUACGGCCAUAAUCGUAUCCGGAUUAGCCGUC--- ..........((((.....-.))))....(((((..(((((.......)))))))))).--- ( -12.40, z-score = -0.92, R) >droYak2.chrX 16246185 57 + 21770863 -AUAUUUUUUGGUUACUGU-AAACCGUUUACGGCCAUAAUCGUAUCCGGAUUAGCCGUC--- -.........((((.....-.))))....(((((..(((((.......)))))))))).--- ( -12.40, z-score = -1.21, R) >droEre2.scaffold_4690 7953345 57 + 18748788 -GUAGUCUUUGGUUACUGU-AAACCGUUUACGGCCAUAAUCGUAUCCGGAUUAGCCGUC--- -.........((((.....-.))))....(((((..(((((.......)))))))))).--- ( -12.40, z-score = -0.80, R) >droAna3.scaffold_13335 287142 51 + 3335858 -----------UUUACUGUUAAACCGUUUACGGCCAUAAUCGUAUCCGGAUUAGCUACCAAC -----------......(((((.(((..(((((......)))))..))).)))))....... ( -13.40, z-score = -3.06, R) >droWil1.scaffold_181096 8300975 59 - 12416693 GCAAGAAAAGAGACACCGGAAAGCCGUUUACGGCCAUAAUCGUAUCCGGAUUAGCCGUC--- ................(((....(((..(((((......)))))..))).....)))..--- ( -13.90, z-score = -1.29, R) >consensus _GUAUUUUUUGGUUACUGU_AAACCGUUUACGGCCAUAAUCGUAUCCGGAUUAGCCGUC___ .......................(((..(((((......)))))..)))............. ( -9.56 = -9.56 + -0.00)
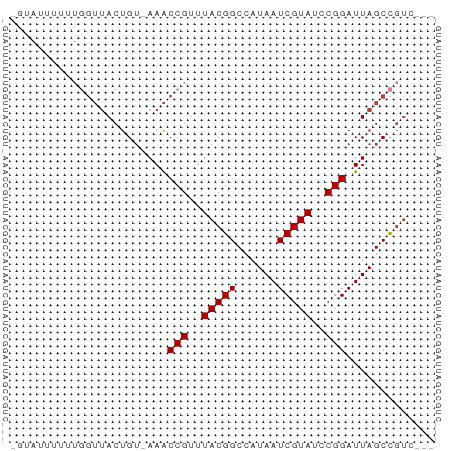
| Location | 17,618,351 – 17,618,409 |
|---|---|
| Length | 58 |
| Sequences | 7 |
| Columns | 62 |
| Reading direction | reverse |
| Mean pairwise identity | 82.64 |
| Shannon entropy | 0.35942 |
| G+C content | 0.43407 |
| Mean single sequence MFE | -14.26 |
| Consensus MFE | -10.62 |
| Energy contribution | -10.73 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.49 |
| SVM RNA-class probability | 0.943749 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
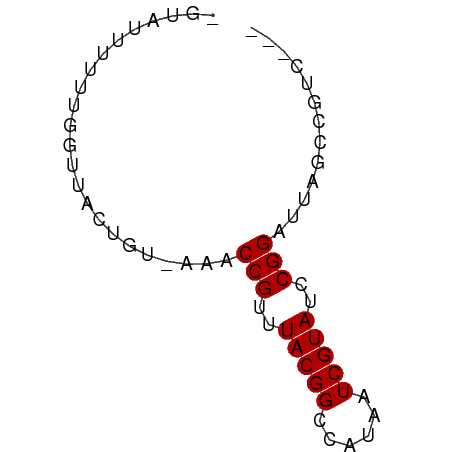
>dm3.chrX 17618351 58 - 22422827 ---GACGGCUAAUCCGGAUACGAUUAUGGCCGUAAACGGUUU-ACAGUAACCAAAAAAUACG ---.(((((((...((....))....)))))))....((((.-.....)))).......... ( -14.60, z-score = -2.16, R) >droSim1.chrX 3680692 58 + 17042790 ---GACGGCUAAUCCGGAUACGAUUAUGGCCGUAAACGGUUU-ACAGUAACCAAAAAAUACA ---.(((((((...((....))....)))))))....((((.-.....)))).......... ( -14.60, z-score = -2.27, R) >droSec1.super_17 1464064 58 - 1527944 ---GACGGCUAAUCCGGAUACGAUUAUGGCCGUAAACGGUUU-ACAGUAACCAAAAAAUACA ---.(((((((...((....))....)))))))....((((.-.....)))).......... ( -14.60, z-score = -2.27, R) >droYak2.chrX 16246185 57 - 21770863 ---GACGGCUAAUCCGGAUACGAUUAUGGCCGUAAACGGUUU-ACAGUAACCAAAAAAUAU- ---.(((((((...((....))....)))))))....((((.-.....)))).........- ( -14.60, z-score = -2.36, R) >droEre2.scaffold_4690 7953345 57 - 18748788 ---GACGGCUAAUCCGGAUACGAUUAUGGCCGUAAACGGUUU-ACAGUAACCAAAGACUAC- ---.(((((((...((....))....)))))))....((((.-.....)))).........- ( -14.60, z-score = -2.11, R) >droAna3.scaffold_13335 287142 51 - 3335858 GUUGGUAGCUAAUCCGGAUACGAUUAUGGCCGUAAACGGUUUAACAGUAAA----------- .......(.(((.(((..((((........))))..))).))).)......----------- ( -9.40, z-score = -0.05, R) >droWil1.scaffold_181096 8300975 59 + 12416693 ---GACGGCUAAUCCGGAUACGAUUAUGGCCGUAAACGGCUUUCCGGUGUCUCUUUUCUUGC ---((.(((....(((((.........(((((....))))).))))).)))))......... ( -17.40, z-score = -1.80, R) >consensus ___GACGGCUAAUCCGGAUACGAUUAUGGCCGUAAACGGUUU_ACAGUAACCAAAAAAUAC_ ....(((((((...((....))....)))))))....((((.......)))).......... (-10.62 = -10.73 + 0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:54:00 2011