
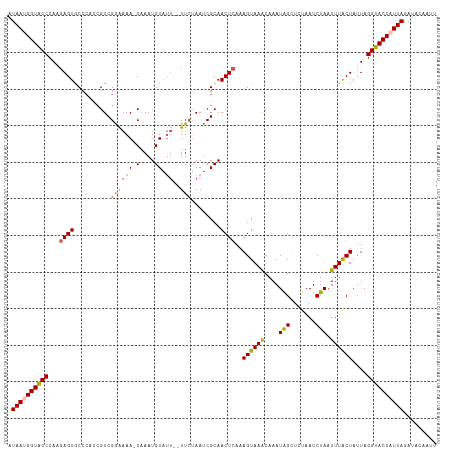
| Sequence ID | dm3.chrX |
|---|---|
| Location | 17,605,607 – 17,605,723 |
| Length | 116 |
| Max. P | 0.899512 |

| Location | 17,605,607 – 17,605,723 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 87.93 |
| Shannon entropy | 0.20768 |
| G+C content | 0.34443 |
| Mean single sequence MFE | -23.36 |
| Consensus MFE | -15.80 |
| Energy contribution | -15.60 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.899512 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 17605607 116 + 22422827 AUAAUGGUACCCAAAAGUGCCCACCGUCGGAAAA-CAAAUGUUGCGAUUAGAACCGCAACUCAAAGUAAACAAAUAGAUCUAAUCUAAUUUACUAUUAGGUACCAUUAAAUACAAUU .((((((((((.....((..((......))...)-)....((((((........))))))....((((((....(((((...))))).))))))....))))))))))......... ( -27.90, z-score = -4.54, R) >droSim1.chrX 3667254 114 - 17042790 AUAAAGGUACCCACGAGUGCCCACCGUCGGAAAA-CAAAUGUAUU--UUCUAAUCGCAACUCAAAGUAAACAAAUAGCUCUAAUCUAAUUUACUACUAGGUACCAUUAGAUACAAUU .....((((((...((((((.....((..(((((-........))--)))..)).)).))))..((((((....(((.......))).))))))....))))))............. ( -19.60, z-score = -1.69, R) >droSec1.super_17 1451214 114 + 1527944 AUAAAGGUACCCAAGAGUGCCCACCGUCGGAAAA-CAAAUGUAUU--UUCUAAUCGCAACUCAAAGUAAACAAAUAGCUCUAAUCUAAUUUGCUACUAGGUACCAUUAGAUACAAUU .....((((((...((((((.....((..(((((-........))--)))..)).)).))))............((((.............))))...))))))............. ( -20.62, z-score = -1.74, R) >droYak2.chrX 16233441 114 + 21770863 AUAAUGGUGCCCAAGAGUACCCACCGUCGAGAAA-CAAAUGUAUU--UUCUAAUCGCAACUCAAAGUAAGCAAAUGGUUGUAAUCUAAUUUACUAUUAGGUACCAUUAGAUACAAAC .((((((((((.((.((((.........((((((-(....)).))--))).....((((((....(....)....)))))).........)))).)).))))))))))......... ( -23.70, z-score = -2.07, R) >droEre2.scaffold_4690 7940724 111 + 18748788 AUAAUGGUACCCAAGAGUACC----GUCGAAAAAGCGAAUGUAUU--UUCUAAUCGCAACUCAAAGUAAGCAAAUGGCUGUAAUCUAAUUUACUAUUAGGUACCAUUAGAUACAAAU .((((((((((...((((..(----(..(((((.((....)).))--)))....))..))))......(((.....)))((((......)))).....))))))))))......... ( -25.00, z-score = -2.93, R) >consensus AUAAUGGUACCCAAGAGUGCCCACCGUCGGAAAA_CAAAUGUAUU__UUCUAAUCGCAACUCAAAGUAAACAAAUAGCUCUAAUCUAAUUUACUAUUAGGUACCAUUAGAUACAAUU .((((((((((...((((........................................))))..((((((....(((.......))).))))))....))))))))))......... (-15.80 = -15.60 + -0.20)


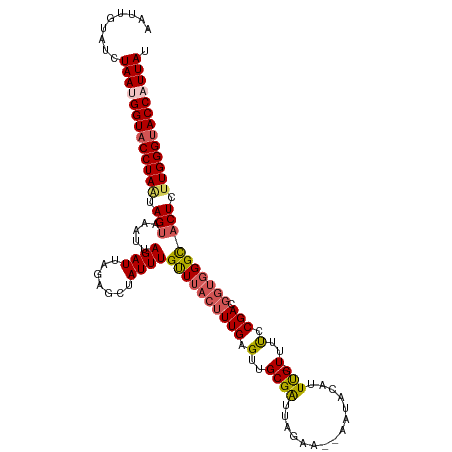
| Location | 17,605,607 – 17,605,723 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 87.93 |
| Shannon entropy | 0.20768 |
| G+C content | 0.34443 |
| Mean single sequence MFE | -29.51 |
| Consensus MFE | -19.56 |
| Energy contribution | -20.52 |
| Covariance contribution | 0.96 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.860384 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 17605607 116 - 22422827 AAUUGUAUUUAAUGGUACCUAAUAGUAAAUUAGAUUAGAUCUAUUUGUUUACUUUGAGUUGCGGUUCUAAUCGCAACAUUUG-UUUUCCGACGGUGGGCACUUUUGGGUACCAUUAU .........(((((((((((((.(((....(((((...)))))...(((((((....((((((((....))))))))....(-((....))))))))))))).))))))))))))). ( -37.70, z-score = -5.14, R) >droSim1.chrX 3667254 114 + 17042790 AAUUGUAUCUAAUGGUACCUAGUAGUAAAUUAGAUUAGAGCUAUUUGUUUACUUUGAGUUGCGAUUAGAA--AAUACAUUUG-UUUUCCGACGGUGGGCACUCGUGGGUACCUUUAU .........(((.((((((((..(((((((.((((.......)))))))))))..(((((.((.(..(((--((((....))-)))))..)...)).).)))).)))))))).))). ( -24.60, z-score = -0.47, R) >droSec1.super_17 1451214 114 - 1527944 AAUUGUAUCUAAUGGUACCUAGUAGCAAAUUAGAUUAGAGCUAUUUGUUUACUUUGAGUUGCGAUUAGAA--AAUACAUUUG-UUUUCCGACGGUGGGCACUCUUGGGUACCUUUAU ....((((((((.(((.((((.(.((((.(((((...((((.....))))..))))).))))(.(..(((--((((....))-)))))..))).)))).))).))))))))...... ( -26.70, z-score = -1.22, R) >droYak2.chrX 16233441 114 - 21770863 GUUUGUAUCUAAUGGUACCUAAUAGUAAAUUAGAUUACAACCAUUUGCUUACUUUGAGUUGCGAUUAGAA--AAUACAUUUG-UUUCUCGACGGUGGGUACUCUUGGGCACCAUUAU .........(((((((.(((((..((((......)))).......(((((((((((((..((((......--.......)))-)..))))).))))))))...))))).))))))). ( -28.92, z-score = -2.34, R) >droEre2.scaffold_4690 7940724 111 - 18748788 AUUUGUAUCUAAUGGUACCUAAUAGUAAAUUAGAUUACAGCCAUUUGCUUACUUUGAGUUGCGAUUAGAA--AAUACAUUCGCUUUUUCGAC----GGUACUCUUGGGUACCAUUAU .........(((((((((((((.((((...........(((.....)))..(.(((((..((((......--.......))))...))))).----).)))).))))))))))))). ( -29.62, z-score = -3.61, R) >consensus AAUUGUAUCUAAUGGUACCUAAUAGUAAAUUAGAUUAGAGCUAUUUGUUUACUUUGAGUUGCGAUUAGAA__AAUACAUUUG_UUUUCCGACGGUGGGCACUCUUGGGUACCAUUAU .........(((((((((((((.(((.....((((.......))))(((((((....((((.((...(((........)))....)).)))))))))))))).))))))))))))). (-19.56 = -20.52 + 0.96)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:53:58 2011