
| Sequence ID | dm3.chrX |
|---|---|
| Location | 17,604,012 – 17,604,205 |
| Length | 193 |
| Max. P | 0.996284 |

| Location | 17,604,012 – 17,604,125 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.08 |
| Shannon entropy | 0.35516 |
| G+C content | 0.45297 |
| Mean single sequence MFE | -35.20 |
| Consensus MFE | -24.76 |
| Energy contribution | -22.92 |
| Covariance contribution | -1.84 |
| Combinations/Pair | 1.39 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.68 |
| SVM RNA-class probability | 0.960271 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 17604012 113 - 22422827 UCGCGCCAAUUGUUGGUGUGAAAAUAAUACCAACGAAAUGCGGUACUUCUGUGUCGGUUGCUGUCUGGGUAACACGCACCUGUAUGGGUUCGCGUCUUGGCUAAGUGUCAUAU------- ....(((((((((((((((.......)))))))))).((((((..((.(.(((.((((((((.....)))))).)))))..)...))..)))))).)))))............------- ( -35.90, z-score = -1.69, R) >droEre2.scaffold_4690 7939282 116 - 18748788 UCGCGCCAAUUGUUGGUAUGAA---AAUACCAACGAAAUGCGGUAUUUCGAAGUCGGUUGCCGUCUGGGGAUGACGCACCUGCUUCGGUUCGUGUCUUAGCUAA-CAUUUUUGAAGUUAC ..((.....((((((((((...---.)))))))))).((((((....(((((((.(((.((.(((.......)))))))).))))))).))))))....))...-............... ( -37.20, z-score = -2.10, R) >droYak2.chrX 16231877 117 - 21770863 UCGCGCCAAUUGUUGGUAUGAA---AAUACCAACGAAAUGUGCUAUUUCUAUAUCGGUUGCUGUCUGGGGAAGGUGCACUUGUUUCGGUUCGUGUCUUAGCUAACCAUUUUUUCAGUUAC ..((((...((((((((((...---.))))))))))...))))............((((((((.(((..((((.....))).)..))).........)))).)))).............. ( -31.90, z-score = -1.90, R) >droSec1.super_17 1449674 113 - 1527944 UCGCGCCAAUUGUUGGUGUGAAAAUAAUACCAACAAAAUGCGGCACUUCUGUGUCGGUUGAUGUCUGGGAUAAACGCAUCUGUGUGGGUUCGCAUCUUCGCUAAAGGUCAUAU------- ..(((((..((((((((((.......)))))))))).....(((((....)))))))).(((((...((((..((((....))))..)))))))))...))............------- ( -35.70, z-score = -2.55, R) >droSim1.chrX 3665687 113 + 17042790 UCGCGCCAAUUGUUGGUGUGAAAAUAAUACCAACAAAAUGCGGCACUUCUGUGUCGGUUGAUGUCUGGGAUAAACGCAUCUGUGUGGGUUCGCGUCUUCGCUAAAUGUCAUAU------- ..(((((..((((((((((.......)))))))))).....(((((....)))))))).(((((...((((..((((....))))..)))))))))...))............------- ( -35.30, z-score = -2.45, R) >consensus UCGCGCCAAUUGUUGGUGUGAAAAUAAUACCAACGAAAUGCGGUACUUCUGUGUCGGUUGCUGUCUGGGAAAAACGCACCUGUGUGGGUUCGCGUCUUAGCUAAACGUCAUAU_______ ..((.(((.((((((((((.......))))))))))....((((((....)))))).........))).....(((((((......)))..))))....))................... (-24.76 = -22.92 + -1.84)

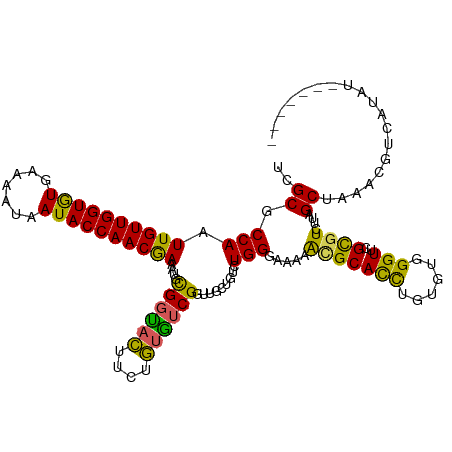

| Location | 17,604,045 – 17,604,165 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.22 |
| Shannon entropy | 0.23823 |
| G+C content | 0.48071 |
| Mean single sequence MFE | -37.21 |
| Consensus MFE | -26.78 |
| Energy contribution | -27.54 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.18 |
| Mean z-score | -3.50 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.91 |
| SVM RNA-class probability | 0.996284 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 17604045 120 + 22422827 GGUGCGUGUUACCCAGACAGCAACCGACACAGAAGUACCGCAUUUCGUUGGUAUUAUUUUCACACCAACAAUUGGCGCGAAAGCCGUGCCAAAGCAGAGAUGCGAUAAAGAAGAGGAGUG (((((.((((.....)))))).))).............((((((((((((((...........))))))..((((((((.....))))))))....))))))))................ ( -38.60, z-score = -3.65, R) >droEre2.scaffold_4690 7939321 116 + 18748788 GGUGCGUCAUCCCCAGACGGCAACCGACUUCGAAAUACCGCAUUUCGUUGGUA---UUUUCAUACCAACAAUUGGCGCGAAAGCCGUGCCAAAGCAGAAAUAAUAA-AAGAAGAGGCGCG .((((.((.((....(.(((....((....)).....))))((((((((((((---(....))))))))..((((((((.....))))))))....))))).....-..)).)).)))). ( -38.50, z-score = -3.43, R) >droYak2.chrX 16231917 117 + 21770863 AGUGCACCUUCCCCAGACAGCAACCGAUAUAGAAAUAGCACAUUUCGUUGGUA---UUUUCAUACCAACAAUUGGCGCGAAAGCCGUGCCAAAGCGGAGAUGCGAUUAAGAAGAGGCGCG .((((..((((........(((.(((.....(((((.....)))))(((((((---(....))))))))..((((((((.....))))))))..)))...)))......))))..)))). ( -40.94, z-score = -4.85, R) >droSec1.super_17 1449707 120 + 1527944 GAUGCGUUUAUCCCAGACAUCAACCGACACAGAAGUGCCGCAUUUUGUUGGUAUUAUUUUCACACCAACAAUUGGCGCGAAAGCCGUGCCAAAGCAGAGAUGCGAUAAAGAAGAGGAGUG (((((..........).))))..((..(((....))).((((((((((((((...........))))))..((((((((.....))))))))....))))))))..........)).... ( -34.00, z-score = -2.78, R) >droSim1.chrX 3665720 120 - 17042790 GAUGCGUUUAUCCCAGACAUCAACCGACACAGAAGUGCCGCAUUUUGUUGGUAUUAUUUUCACACCAACAAUUGGCGCGAAAGCCGUGCCAAAGCAGAGAUGCGAUAAAGAAGAGGAGUG (((((..........).))))..((..(((....))).((((((((((((((...........))))))..((((((((.....))))))))....))))))))..........)).... ( -34.00, z-score = -2.78, R) >consensus GGUGCGUCUUCCCCAGACAGCAACCGACACAGAAGUACCGCAUUUCGUUGGUAUUAUUUUCACACCAACAAUUGGCGCGAAAGCCGUGCCAAAGCAGAGAUGCGAUAAAGAAGAGGAGUG ...((((((.....)))).....((..(((....))).((((((((((((((...........))))))..((((((((.....))))))))....))))))))..........)).)). (-26.78 = -27.54 + 0.76)

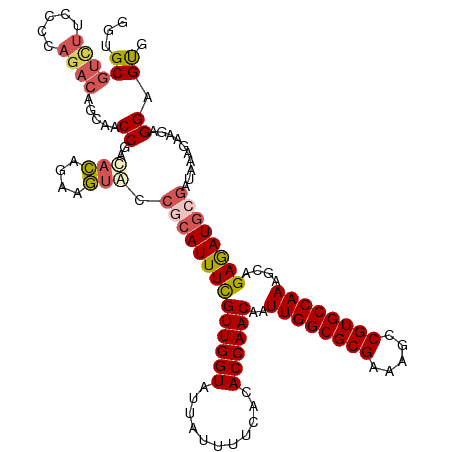
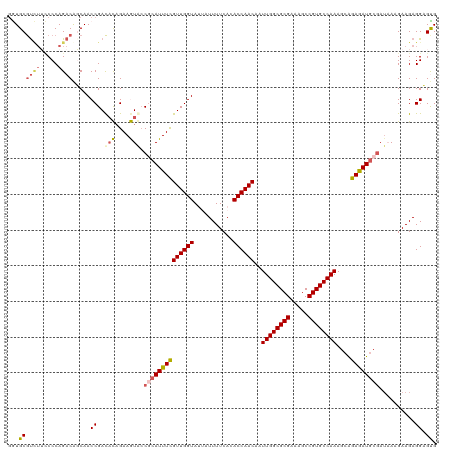
| Location | 17,604,045 – 17,604,165 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.22 |
| Shannon entropy | 0.23823 |
| G+C content | 0.48071 |
| Mean single sequence MFE | -40.12 |
| Consensus MFE | -26.22 |
| Energy contribution | -25.58 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.24 |
| Mean z-score | -3.66 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.83 |
| SVM RNA-class probability | 0.995687 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 17604045 120 - 22422827 CACUCCUCUUCUUUAUCGCAUCUCUGCUUUGGCACGGCUUUCGCGCCAAUUGUUGGUGUGAAAAUAAUACCAACGAAAUGCGGUACUUCUGUGUCGGUUGCUGUCUGGGUAACACGCACC .............((((((((.((....(((((.((.....)).)))))..((((((((.......)))))))))).)))))))).....(((.((((((((.....)))))).))))). ( -39.70, z-score = -3.69, R) >droEre2.scaffold_4690 7939321 116 - 18748788 CGCGCCUCUUCUU-UUAUUAUUUCUGCUUUGGCACGGCUUUCGCGCCAAUUGUUGGUAUGAAAA---UACCAACGAAAUGCGGUAUUUCGAAGUCGGUUGCCGUCUGGGGAUGACGCACC .((((.(((((..-.....(((((....(((((.((.....)).)))))..((((((((....)---))))))))))))((((((..(((....))).))))))..))))).).)))... ( -41.70, z-score = -3.17, R) >droYak2.chrX 16231917 117 - 21770863 CGCGCCUCUUCUUAAUCGCAUCUCCGCUUUGGCACGGCUUUCGCGCCAAUUGUUGGUAUGAAAA---UACCAACGAAAUGUGCUAUUUCUAUAUCGGUUGCUGUCUGGGGAAGGUGCACU .((((((..(((..((.(((...(((...(((((((((......)))..((((((((((....)---)))))))))...)))))).........))).))).))..)))..))))))... ( -46.00, z-score = -5.07, R) >droSec1.super_17 1449707 120 - 1527944 CACUCCUCUUCUUUAUCGCAUCUCUGCUUUGGCACGGCUUUCGCGCCAAUUGUUGGUGUGAAAAUAAUACCAACAAAAUGCGGCACUUCUGUGUCGGUUGAUGUCUGGGAUAAACGCAUC ...........(((((((((((...((.(((((.((.....)).)))))((((((((((.......))))))))))....((((((....)))))))).)))))....))))))...... ( -36.60, z-score = -3.18, R) >droSim1.chrX 3665720 120 + 17042790 CACUCCUCUUCUUUAUCGCAUCUCUGCUUUGGCACGGCUUUCGCGCCAAUUGUUGGUGUGAAAAUAAUACCAACAAAAUGCGGCACUUCUGUGUCGGUUGAUGUCUGGGAUAAACGCAUC ...........(((((((((((...((.(((((.((.....)).)))))((((((((((.......))))))))))....((((((....)))))))).)))))....))))))...... ( -36.60, z-score = -3.18, R) >consensus CACUCCUCUUCUUUAUCGCAUCUCUGCUUUGGCACGGCUUUCGCGCCAAUUGUUGGUGUGAAAAUAAUACCAACGAAAUGCGGUACUUCUGUGUCGGUUGCUGUCUGGGAAAAACGCACC ....(((..........((((.......(((((.((.....)).)))))((((((((((.......)))))))))).))))(((((....)))))...........)))........... (-26.22 = -25.58 + -0.64)

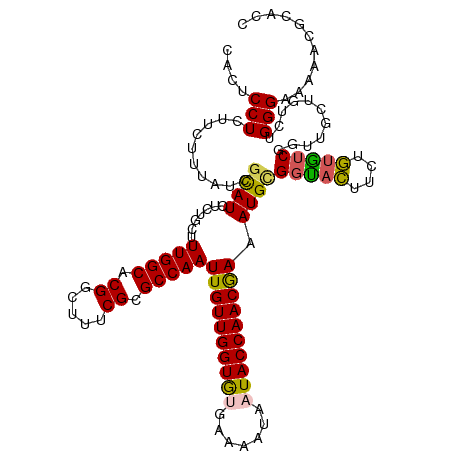
| Location | 17,604,085 – 17,604,205 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.28 |
| Shannon entropy | 0.16327 |
| G+C content | 0.49083 |
| Mean single sequence MFE | -36.48 |
| Consensus MFE | -30.50 |
| Energy contribution | -30.90 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.84 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.81 |
| SVM RNA-class probability | 0.969312 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 17604085 120 + 22422827 CAUUUCGUUGGUAUUAUUUUCACACCAACAAUUGGCGCGAAAGCCGUGCCAAAGCAGAGAUGCGAUAAAGAAGAGGAGUGCAGAGAGGCGGCAGUCUGUGGAGAAACACCAGUGGAAUCG .(((((((((((.((.(((((((........((((((((.....)))))))).(((....)))...........(((.(((.(.....).))).)))))))))))).))))))))))).. ( -36.60, z-score = -2.40, R) >droEre2.scaffold_4690 7939361 113 + 18748788 CAUUUCGUUGGUAUU---UUCAUACCAACAAUUGGCGCGAAAGCCGUGCCAAAGCAGAAAUAAUA-AAAGAAGAGGCGCGCAGAGAG---GCAGUCUGUGGAGAAACACCAGUGGAAUCG .(((((((((((((.---...))))))))..((((((((.....))))))))....)))))....-......((..(.((((((...---....))))))......((....)))..)). ( -33.20, z-score = -2.61, R) >droYak2.chrX 16231957 112 + 21770863 CAUUUCGUUGGUAUU---UUCAUACCAACAAUUGGCGCGAAAGCCGUGCCAAAGCGGAGAUGCGAUUAAGAAGAGGCGCGCAGAG-----GCAGUCUGUGGAGAAACACCAGCGGAGUCG .(((((((((((.((---(((...((.....((((((((.....)))))))).(((....)))...........))..((((((.-----....)))))))))))..))))))))))).. ( -39.00, z-score = -2.57, R) >droSec1.super_17 1449747 120 + 1527944 CAUUUUGUUGGUAUUAUUUUCACACCAACAAUUGGCGCGAAAGCCGUGCCAAAGCAGAGAUGCGAUAAAGAAGAGGAGUGCAGAGAGGCGGCAGUCUGCGGAGAAACACCAGUGGAAUCA .((((..(((((.((.(((((((.((.....((((((((.....)))))))).(((....)))...........)).))(((((..(....)..)))))))))))).)))))..)))).. ( -36.80, z-score = -2.51, R) >droSim1.chrX 3665760 120 - 17042790 CAUUUUGUUGGUAUUAUUUUCACACCAACAAUUGGCGCGAAAGCCGUGCCAAAGCAGAGAUGCGAUAAAGAAGAGGAGUGCAGAGAGGCGGCAGUCUGCGGAGAAACACCAGUGGAAUCA .((((..(((((.((.(((((((.((.....((((((((.....)))))))).(((....)))...........)).))(((((..(....)..)))))))))))).)))))..)))).. ( -36.80, z-score = -2.51, R) >consensus CAUUUCGUUGGUAUUAUUUUCACACCAACAAUUGGCGCGAAAGCCGUGCCAAAGCAGAGAUGCGAUAAAGAAGAGGAGUGCAGAGAGGCGGCAGUCUGUGGAGAAACACCAGUGGAAUCG .(((((((((((....(((((...((.....((((((((.....)))))))).(((....)))...........))..((((((..........)))))))))))..))))))))))).. (-30.50 = -30.90 + 0.40)


| Location | 17,604,085 – 17,604,205 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.28 |
| Shannon entropy | 0.16327 |
| G+C content | 0.49083 |
| Mean single sequence MFE | -30.28 |
| Consensus MFE | -24.75 |
| Energy contribution | -23.59 |
| Covariance contribution | -1.16 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.82 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.48 |
| SVM RNA-class probability | 0.942049 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 17604085 120 - 22422827 CGAUUCCACUGGUGUUUCUCCACAGACUGCCGCCUCUCUGCACUCCUCUUCUUUAUCGCAUCUCUGCUUUGGCACGGCUUUCGCGCCAAUUGUUGGUGUGAAAAUAAUACCAACGAAAUG ...(((...(((((((............((((......(((................)))....(((....)))))))((((((((((.....))))))))))..)))))))..)))... ( -28.09, z-score = -1.65, R) >droEre2.scaffold_4690 7939361 113 - 18748788 CGAUUCCACUGGUGUUUCUCCACAGACUGC---CUCUCUGCGCGCCUCUUCUUU-UAUUAUUUCUGCUUUGGCACGGCUUUCGCGCCAAUUGUUGGUAUGAA---AAUACCAACGAAAUG ..........(((((.(((....)))..((---......)))))))........-...((((((....(((((.((.....)).)))))..((((((((...---.)))))))))))))) ( -29.30, z-score = -2.66, R) >droYak2.chrX 16231957 112 - 21770863 CGACUCCGCUGGUGUUUCUCCACAGACUGC-----CUCUGCGCGCCUCUUCUUAAUCGCAUCUCCGCUUUGGCACGGCUUUCGCGCCAAUUGUUGGUAUGAA---AAUACCAACGAAAUG .((..((((.(((((((......)))).))-----)...))).)..)).........((......)).(((((.((.....)).)))))((((((((((...---.)))))))))).... ( -31.60, z-score = -2.14, R) >droSec1.super_17 1449747 120 - 1527944 UGAUUCCACUGGUGUUUCUCCGCAGACUGCCGCCUCUCUGCACUCCUCUUCUUUAUCGCAUCUCUGCUUUGGCACGGCUUUCGCGCCAAUUGUUGGUGUGAAAAUAAUACCAACAAAAUG .........(((((((.....(((((..(....)..)))))..............(((((((...((.(((((.((.....)).)))))..)).)))))))....)))))))........ ( -31.20, z-score = -2.76, R) >droSim1.chrX 3665760 120 + 17042790 UGAUUCCACUGGUGUUUCUCCGCAGACUGCCGCCUCUCUGCACUCCUCUUCUUUAUCGCAUCUCUGCUUUGGCACGGCUUUCGCGCCAAUUGUUGGUGUGAAAAUAAUACCAACAAAAUG .........(((((((.....(((((..(....)..)))))..............(((((((...((.(((((.((.....)).)))))..)).)))))))....)))))))........ ( -31.20, z-score = -2.76, R) >consensus CGAUUCCACUGGUGUUUCUCCACAGACUGCCGCCUCUCUGCACUCCUCUUCUUUAUCGCAUCUCUGCUUUGGCACGGCUUUCGCGCCAAUUGUUGGUGUGAAAAUAAUACCAACGAAAUG ......((..(((((........(((.(((.........)))....)))........)))))..))..(((((.((.....)).)))))((((((((((.......)))))))))).... (-24.75 = -23.59 + -1.16)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:53:56 2011