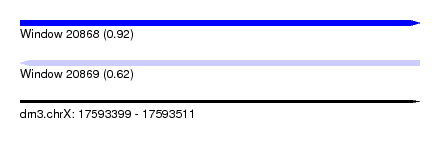
| Sequence ID | dm3.chrX |
|---|---|
| Location | 17,593,399 – 17,593,511 |
| Length | 112 |
| Max. P | 0.920992 |

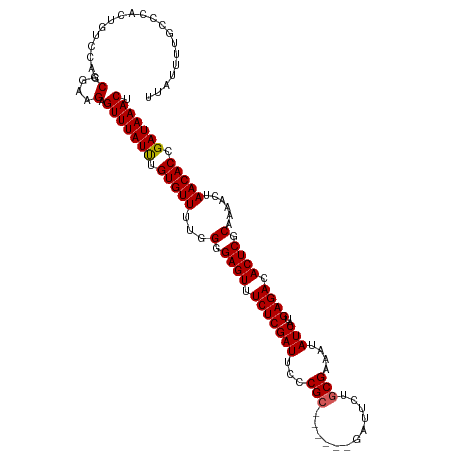
| Location | 17,593,399 – 17,593,511 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 95.80 |
| Shannon entropy | 0.07152 |
| G+C content | 0.43364 |
| Mean single sequence MFE | -33.36 |
| Consensus MFE | -30.42 |
| Energy contribution | -30.26 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.91 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.28 |
| SVM RNA-class probability | 0.920992 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 17593399 112 + 22422827 AGUUUAUCGGUGUUAGUUUUGCGAGUGUCUCAUGAUAUUUCGCAGAAUC------GCGGGAAUCGAGAAACUCCCCAAAACACAAAUAAACUCUUCCGGUGGACAGUGGGCAAAAUAA (((((((..(((((.((((((((((((((....)))).)))))))))).------(.((((..........)))))..)))))..)))))))..((((.(....).))))........ ( -31.50, z-score = -1.87, R) >droEre2.scaffold_4690 7928547 118 + 18748788 AGUUUAUCGGUGUUAGUUUUGCGAGUGUCUCAUGAUAUUUCGCAGAAUCAGAAUCGCGGGAAUCGAGAAACUCCCCAAAACACAGAUAAACUCUUCCGGUGGACAGUGGGCAAAAUAA ((((((((.(((((.((((((((((((((....)))).)))))))))).......(.((((..........)))))..))))).))))))))..((((.(....).))))........ ( -34.10, z-score = -2.01, R) >droYak2.chrX 16221280 111 + 21770863 UGUUUAUCGGUGUUAGUUUUGCGAGUGUCUCAUGAUAUUUCGUAGAAUC------GCGGGAAUCGAGAAACUCCCCAAAACACAGAUAAACUCUUCCGGUGGACAGUGGGCAAAAUA- .(((((((.(((((.((((((((((((((....)))).)))))))))).------(.((((..........)))))..))))).)))))))...((((.(....).)))).......- ( -31.60, z-score = -1.66, R) >droSec1.super_17 1439211 112 + 1527944 AGUUUAUCGGUGUUGGUUUUGCGAGUGUCUCAUGAUAUUUCGCAGAAUC------GCGGGAAUCGAGAAACUCCCCAAAACACAAAUAAACUCUUCCGGUGGACAGUGGGCAAAAUAA (((((((..((((((((((((((((((((....)))).)))))))))))------(.((((..........)))))..)))))..)))))))..((((.(....).))))........ ( -34.80, z-score = -2.46, R) >droSim1.chrX 3655153 112 - 17042790 AGUUUAUCGGUGUUGGUUUUGCGAGUGUCUCAUGAUAUUUCGCAGAAUC------GCGGGAAUCGAGAAACUCCCCAAAACACAAAUAAACUCUUCCGGUGGACAGUGGGCAAAAUAA (((((((..((((((((((((((((((((....)))).)))))))))))------(.((((..........)))))..)))))..)))))))..((((.(....).))))........ ( -34.80, z-score = -2.46, R) >consensus AGUUUAUCGGUGUUAGUUUUGCGAGUGUCUCAUGAUAUUUCGCAGAAUC______GCGGGAAUCGAGAAACUCCCCAAAACACAAAUAAACUCUUCCGGUGGACAGUGGGCAAAAUAA .(((((((((.(((.((((((((((((((....)))).)))))))))).......(.((((..........)))))............)))....))))))))).............. (-30.42 = -30.26 + -0.16)

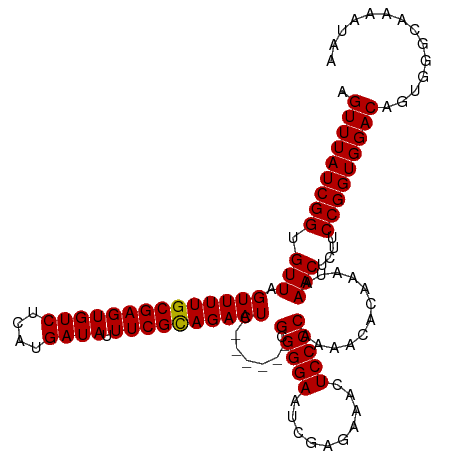
| Location | 17,593,399 – 17,593,511 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 95.80 |
| Shannon entropy | 0.07152 |
| G+C content | 0.43364 |
| Mean single sequence MFE | -28.02 |
| Consensus MFE | -24.80 |
| Energy contribution | -24.76 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.89 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.624807 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 17593399 112 - 22422827 UUAUUUUGCCCACUGUCCACCGGAAGAGUUUAUUUGUGUUUUGGGGAGUUUCUCGAUUCCCGC------GAUUCUGCGAAAUAUCAUGAGACACUCGCAAAACUAACACCGAUAAACU ....................(....)((((((((.(((((...(.((((.(((((((...(((------(....))))....)))..)))).)))).)......))))).)))))))) ( -27.90, z-score = -1.65, R) >droEre2.scaffold_4690 7928547 118 - 18748788 UUAUUUUGCCCACUGUCCACCGGAAGAGUUUAUCUGUGUUUUGGGGAGUUUCUCGAUUCCCGCGAUUCUGAUUCUGCGAAAUAUCAUGAGACACUCGCAAAACUAACACCGAUAAACU ....................(....)((((((((.(((((...(.((((.(((((((...(((((.......)).)))....)))..)))).)))).)......))))).)))))))) ( -29.60, z-score = -1.66, R) >droYak2.chrX 16221280 111 - 21770863 -UAUUUUGCCCACUGUCCACCGGAAGAGUUUAUCUGUGUUUUGGGGAGUUUCUCGAUUCCCGC------GAUUCUACGAAAUAUCAUGAGACACUCGCAAAACUAACACCGAUAAACA -...................(....).(((((((.(((((..((((((((....)))))))((------((.(((..((....))...)))...))))....).))))).))))))). ( -28.20, z-score = -1.99, R) >droSec1.super_17 1439211 112 - 1527944 UUAUUUUGCCCACUGUCCACCGGAAGAGUUUAUUUGUGUUUUGGGGAGUUUCUCGAUUCCCGC------GAUUCUGCGAAAUAUCAUGAGACACUCGCAAAACCAACACCGAUAAACU ....................(....)((((((((.(((((...(.((((.(((((((...(((------(....))))....)))..)))).)))).)......))))).)))))))) ( -27.20, z-score = -1.34, R) >droSim1.chrX 3655153 112 + 17042790 UUAUUUUGCCCACUGUCCACCGGAAGAGUUUAUUUGUGUUUUGGGGAGUUUCUCGAUUCCCGC------GAUUCUGCGAAAUAUCAUGAGACACUCGCAAAACCAACACCGAUAAACU ....................(....)((((((((.(((((...(.((((.(((((((...(((------(....))))....)))..)))).)))).)......))))).)))))))) ( -27.20, z-score = -1.34, R) >consensus UUAUUUUGCCCACUGUCCACCGGAAGAGUUUAUUUGUGUUUUGGGGAGUUUCUCGAUUCCCGC______GAUUCUGCGAAAUAUCAUGAGACACUCGCAAAACUAACACCGAUAAACU ....................(....).(((((((.(((((...(((((((....))))))).............(((((....((....))...))))).....))))).))))))). (-24.80 = -24.76 + -0.04)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:53:51 2011