
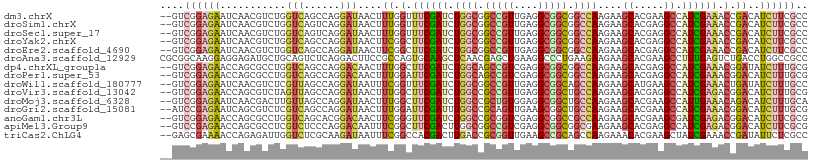
| Sequence ID | dm3.chrX |
|---|---|
| Location | 17,556,517 – 17,556,623 |
| Length | 106 |
| Max. P | 0.640879 |

| Location | 17,556,517 – 17,556,623 |
|---|---|
| Length | 106 |
| Sequences | 15 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 80.53 |
| Shannon entropy | 0.45205 |
| G+C content | 0.58226 |
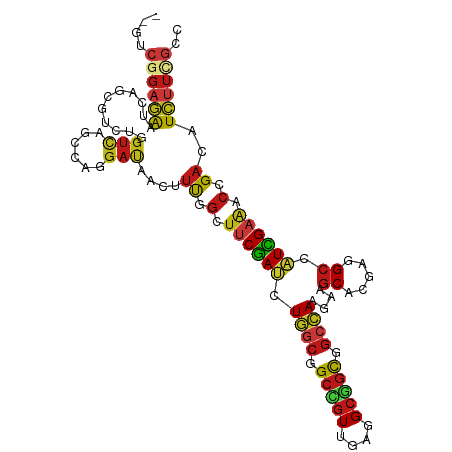
| Mean single sequence MFE | -39.81 |
| Consensus MFE | -21.17 |
| Energy contribution | -21.34 |
| Covariance contribution | 0.17 |
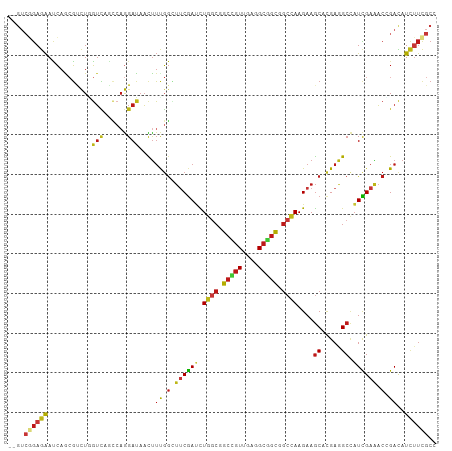
| Combinations/Pair | 1.48 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.640879 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 17556517 106 + 22422827 --GUCGGAGAAUCAACGUCUGGUCAGCCAGGAUAACUUUGGUUUCGAUCUGGCGGCCGUUGAGGCGGCGGCCAAGAAGCACGAAGCCAUCGAAACCGACAUCUUCGCC --..((((((.......(((((....)))))......((((((((((((((((.(((((....))))).))).))).((.....))..))))))))))..)))))).. ( -43.90, z-score = -2.87, R) >droSim1.chrX 3630965 106 - 17042790 --GUCGGAGAAUCAACGUCUGGUCAGUCAGGAUAACUUUGGUUUCGAUCUGGCGGCCGUUGAGGCGGCGGCCAAGAAGCACGAGGCCAUCGAAACCGACAUCUUCGCC --..((((((.......(((((....)))))......((((((((((((((((.(((((....))))).))).))).((.....))..))))))))))..)))))).. ( -40.20, z-score = -1.71, R) >droSec1.super_17 1399141 106 + 1527944 --GUCGGAGAAUCAACGUCUGGUCAGUCAGGAUAACUUUGGUUUCGAUCUGGCGGCCGUUGAGGCGGCGGCCAAGAAGCACGAGGCCAUCGAAACCGACAUCUUCGCC --..((((((.......(((((....)))))......((((((((((((((((.(((((....))))).))).))).((.....))..))))))))))..)))))).. ( -40.20, z-score = -1.71, R) >droYak2.chrX 16183873 106 + 21770863 --GUCGGAGAAUCAACGUCUGGUCAGCCAGGAUAACUUCGGCUUCGAUCUGGCGGCCGUUGAGGCGGCGGCCAAGAAGCACGAGGCCAUCGAAACCGACAUCUUCGCC --(((((...........((((....))))(((..((((((((((....((((.(((((....))))).)))).))))).)))))..)))....)))))......... ( -47.50, z-score = -3.28, R) >droEre2.scaffold_4690 7892983 106 + 18748788 --GUCGGAGAAUCAACGUCUGGUCAGCCAGGAUAACUUCGGCUUCGAUCUGGCGGCCGUUGAGGCGGCGGCCAAGAAGCACGAGGCCAUCGAAACCGACAUCUUCGCC --(((((...........((((....))))(((..((((((((((....((((.(((((....))))).)))).))))).)))))..)))....)))))......... ( -47.50, z-score = -3.28, R) >droAna3.scaffold_12929 1362097 108 - 3277472 CGCGGCAAGGAGGAGAUGCUGCAGUCUCAGGACUUCCGCCAGUGCAAGCUCAACGAGCUGAAGGCCCUGAAGAAGAAGCACGAAGCCUUUGAGUCUGACCUGGCCGCC .(((((.(((.(((..((((....(((((((.((((.((....)).(((((...)))))))))..)))).)))...))))(((.....)))..)))..))).))))). ( -38.50, z-score = -0.19, R) >dp4.chrXL_group1a 1534598 106 + 9151740 --GUCGGAGAACCAGCGCCUGGUCAGCCAGGACAACUUUGGCUUCGAUCUGGCAGCCGUCGAGGCGGCGGCCAAGAAGCACGAGGCCAUCGAAACGGAUAUCUUUGCG --(.((((((.((...((((((....))))).)..(..((((((((.((((((.(((((....))))).))).)))....))))))))..)....))...))))))). ( -45.00, z-score = -2.32, R) >droPer1.super_53 257423 106 - 525408 --GUCGGAGAACCAGCGCCUGGUCAGCCAGGACAACUUUGGAUUCGAUCUGGCAGCCGUCGAGGCGGCGGCCAAGAAGCACGAGGCCAUCGAAACGGACAUCUUUGCG --(.((((((.((...((((((....))))).)..(..(((.((((.((((((.(((((....))))).))).)))....)))).)))..)....))...))))))). ( -38.40, z-score = -0.66, R) >droWil1.scaffold_180777 2122157 106 + 4753960 --GUCGGAGAAUCAACGUCUCGUUAGCCAGGAUAAUUUCGGUUUCGAUCUGGCGGCCGUUGAGGCGGCAGCCAAGAAGCAUGAAGCCAUCGAAACUGAUAUCUUUGCC --....((((.......))))....((.((((((...((((((((((((((((.(((((....))))).))).))).((.....))..)))))))))))))))).)). ( -39.10, z-score = -2.46, R) >droVir3.scaffold_13042 2109805 106 - 5191987 --GUCGGAGAACCAGCGUCUAGUUAGCCAGGAUAACUUUGGCUUCGAUCUGGCCGCCGUUGAGGCGGCUGCCAAGAAGCACGAGGCCAUCGAGACGGACAUCUUUGCG --(((((....))..((((((((((.......))))).((((((((....(((((((.....)))))))((......)).))))))))...))))))))......... ( -41.50, z-score = -1.86, R) >droMoj3.scaffold_6328 585248 106 + 4453435 --GUCGGAGAAUCAACGACUUGUUAGCCAGGAUAACUUUGGCUUCGAUCUGGCCGCUGUGGAGGCGGCUGCCAAGAAGCACGAAGCCAUUGAAACAGACAUCUUUGCA --((((.........))))......((.(((((..((.((((((((....(((((((.....)))))))((......)).)))))))).......))..))))).)). ( -35.20, z-score = -1.36, R) >droGri2.scaffold_15081 3157791 106 + 4274704 --AUCCGAGAAUCAGCGUCUCGUCAGCCAGGAUAACUUUGGAUUCGAUUUGGCCGCAGUUGAAGCGGCUGCCAAGAAGCACGAAGCCAUCGAAACGGACAUCUUUGCG --...(((((.......)))))...((.(((((..(((((..(((.....((((((.......)))))).....)))...)))))((........))..))))).)). ( -28.60, z-score = 0.56, R) >anoGam1.chr3L 11671702 106 + 41284009 --GUCGGAGAACCAGCGCCUGGUCAGCACGGACAACUUCGGGUUCGAUCUGGCCGCGGUCGAGGCGGCCGCCAAGAAGCACGAAGCGAUCGAGACGGACAUCUUCGCG --(.((((((.((.((..((((((((..(((((..(....))))))..))))))(((((((...)))))))..))..)).(((.....)))....))...))))))). ( -38.00, z-score = 0.62, R) >apiMel3.Group9 9404471 106 - 10282195 --GUCCGAGAACCAGCGCCUCGUCUCCCAGGACAAUUUCGGCUUCGACUUGGCGGCCGUCGAGGCGGCGGCGAAGAAGCACGAGGCCAUCGAGACGGACAUCUUCGCG --(((((((.........)))(((((...(((....)))(((((((.(((.((.(((((....))))).)).))).....)))))))...)))))))))......... ( -44.20, z-score = -0.99, R) >triCas2.ChLG4 4743265 106 - 13894384 --GAGCGAAAACCAGAGAUUGGUCUCGCAAGAUAAUUUCGGCCACGACUUGACCGCGGUUGAAGCCGCAGCCAAGAAACACGAAGCUAUCGAAACCGAUAUUCUCGCC --..((((..(((((...))))).))))..((.(((.((((......((((.(.(((((....))))).).)))).....(((.....)))...)))).))).))... ( -29.40, z-score = -1.50, R) >consensus __GUCGGAGAAUCAGCGUCUGGUCAGCCAGGAUAACUUUGGCUUCGAUCUGGCGGCCGUUGAGGCGGCGGCCAAGAAGCACGAGGCCAUCGAAACCGACAUCUUCGCC ....((((((...........(((......)))....((.(.((((((.((((.(((((....))))).))))....((.....)).)))))).).))..)))))).. (-21.17 = -21.34 + 0.17)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:53:45 2011