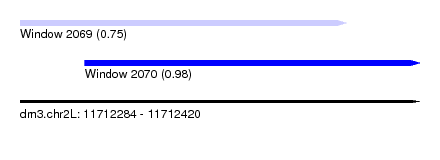
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 11,712,284 – 11,712,420 |
| Length | 136 |
| Max. P | 0.977887 |

| Location | 11,712,284 – 11,712,395 |
|---|---|
| Length | 111 |
| Sequences | 7 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 84.49 |
| Shannon entropy | 0.31030 |
| G+C content | 0.46589 |
| Mean single sequence MFE | -37.49 |
| Consensus MFE | -23.20 |
| Energy contribution | -26.13 |
| Covariance contribution | 2.92 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.747903 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
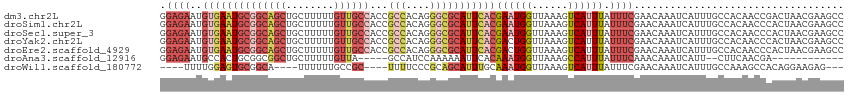
>dm3.chr2L 11712284 111 + 23011544 AGCUGUAGUGCUGCGGUUGAUGGGAGAAUGUGAAUGCGGCAGCUGCUUUUUGUUGCCACCGCCACAGGGCGCAUUCACGAAUGGUUAAAGUCAUUUAUUUCGAACAAAUCA (((((((....)))))))(((..(.(((.((((((((((((((........))))))...(((....)))))))))))((((((......))))))..)))...)..))). ( -39.00, z-score = -1.91, R) >droSim1.chr2L 11521759 111 + 22036055 AGCUGUAGUGCUGCGGUUGAUGGGAGAAUGUGAAUGCGGCAGCUGCUUUUUGUUGCCACCGCCACAGGGCGCAUUCACGAAUGGUUAAAGUCAUUUAUUUCGAACAAAUCA (((((((....)))))))(((..(.(((.((((((((((((((........))))))...(((....)))))))))))((((((......))))))..)))...)..))). ( -39.00, z-score = -1.91, R) >droSec1.super_3 7109792 111 + 7220098 AGCUGUAGUGCUGCGGUUGAUGGGAGAAUGUGAAUGCGGCAGCUGCUUUUUGUUGCCACCGCCACAGGGCGCAUUCACGAAUGGUUAAAGUCAUUUAUUUCGAACAAAUCA (((((((....)))))))(((..(.(((.((((((((((((((........))))))...(((....)))))))))))((((((......))))))..)))...)..))). ( -39.00, z-score = -1.91, R) >droYak2.chr2L 8136838 111 + 22324452 CGCUGUAGUGCUGCGGUUGAUGGGAGAAUGUGAAUGCGGCAGCUGCUUUUUGUUGCCACCGCCACAGGGCGCAUUCACGACUGGUUAAAGUCAUUUAUUUCGAACAAAUCA .((((((....))))))((((..(.(((.((((((((((((((........))))))...(((....)))))))))))((((......))))......)))...)..)))) ( -40.50, z-score = -2.15, R) >droEre2.scaffold_4929 12943386 111 - 26641161 AGCUGUAGUGCUGCGGUUGAUGGGAGAAUGUGAAUGCGGCAGCUGCUUUUUGUUGCCACCGCCACAGGGCGCAUUCACGACUGGUUAAAGUCAUUUAUUUCGAACAAAUCA (((((((....)))))))(((..(.(((.((((((((((((((........))))))...(((....)))))))))))((((......))))......)))...)..))). ( -41.30, z-score = -2.46, R) >droAna3.scaffold_12916 9428193 106 - 16180835 AGCUGAAGUACUGCGGUUUCAUGGAGAAUGCCACUGCGGCGGCUGCUUUUUGUUA-----GCCAUCCAAAAAAUUCACAAAUGGUUAAAGCCAUUUAUUUCAAACAAAUCA ...((((((.(((((((..(((.....)))..))))))).((((((.....).))-----)))...............(((((((....)))))))))))))......... ( -27.00, z-score = -0.98, R) >droWil1.scaffold_180772 5180635 102 - 8906247 AGCUGAAGUGCUGCGGUAUGCUGCCUUUUG-GAGUGCGGCA----UUUUUUGCCGC----UUUUCCCGCAGCAUUUGCAAAUGGUUAAAGUCAUUUAUUUCGAACAAAUCA .((..(((((((((((...(((.((....)-))))((((((----.....))))))----.....)))))))))))))((((((......))))))............... ( -36.60, z-score = -2.91, R) >consensus AGCUGUAGUGCUGCGGUUGAUGGGAGAAUGUGAAUGCGGCAGCUGCUUUUUGUUGCCACCGCCACAGGGCGCAUUCACGAAUGGUUAAAGUCAUUUAUUUCGAACAAAUCA (((((((....))))))).....((((..((((((((((((((........))))))...(((....)))))))))))((((((......)))))).)))).......... (-23.20 = -26.13 + 2.92)
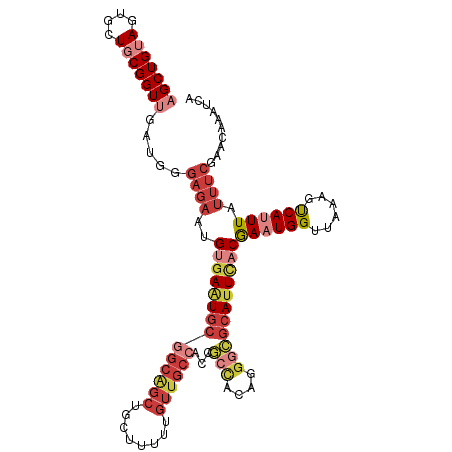
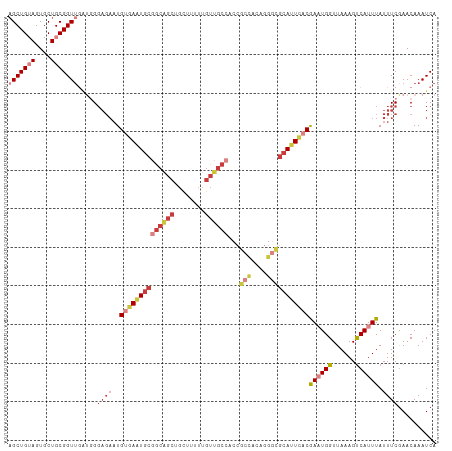
| Location | 11,712,306 – 11,712,420 |
|---|---|
| Length | 114 |
| Sequences | 7 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 80.18 |
| Shannon entropy | 0.40125 |
| G+C content | 0.45565 |
| Mean single sequence MFE | -32.60 |
| Consensus MFE | -18.88 |
| Energy contribution | -21.37 |
| Covariance contribution | 2.50 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.98 |
| SVM RNA-class probability | 0.977887 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
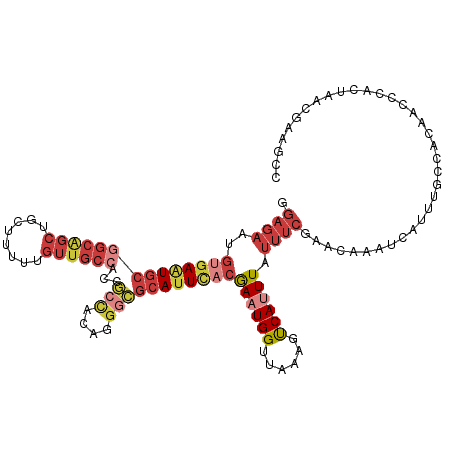
>dm3.chr2L 11712306 114 + 23011544 GGAGAAUGUGAAUGCGGCAGCUGCUUUUUGUUGCCACCGCCACAGGGCGCAUUCACGAAUGGUUAAAGUCAUUUAUUUCGAACAAAUCAUUUGCCACAACCGACUAACGAAGCC ((.....((((((((((((((........))))))...(((....)))))))))))((((((((....((.........))...)))))))).......))............. ( -34.30, z-score = -2.17, R) >droSim1.chr2L 11521781 114 + 22036055 GGAGAAUGUGAAUGCGGCAGCUGCUUUUUGUUGCCACCGCCACAGGGCGCAUUCACGAAUGGUUAAAGUCAUUUAUUUCGAACAAAUCAUUUGCCACAACCCACUAACGAAGCC ((.....((((((((((((((........))))))...(((....)))))))))))((((((((....((.........))...))))))))........))............ ( -34.10, z-score = -2.22, R) >droSec1.super_3 7109814 114 + 7220098 GGAGAAUGUGAAUGCGGCAGCUGCUUUUUGUUGCCACCGCCACAGGGCGCAUUCACGAAUGGUUAAAGUCAUUUAUUUCGAACAAAUCAUUUGCCACAACCCACUAACGAAGCC ((.....((((((((((((((........))))))...(((....)))))))))))((((((((....((.........))...))))))))........))............ ( -34.10, z-score = -2.22, R) >droYak2.chr2L 8136860 114 + 22324452 GGAGAAUGUGAAUGCGGCAGCUGCUUUUUGUUGCCACCGCCACAGGGCGCAUUCACGACUGGUUAAAGUCAUUUAUUUCGAACAAAUCAUUUGCCACAACCCACUAACGAAGCC ((..(((((((((((((((((........))))))...(((....)))))))))))((((......))))..................)))..))................... ( -36.00, z-score = -2.80, R) >droEre2.scaffold_4929 12943408 114 - 26641161 GGAGAAUGUGAAUGCGGCAGCUGCUUUUUGUUGCCACCGCCACAGGGCGCAUUCACGACUGGUUAAAGUCAUUUAUUUCGAACAAAUCAUUUGCCACAACCCACUAACGAAGCC ((..(((((((((((((((((........))))))...(((....)))))))))))((((......))))..................)))..))................... ( -36.00, z-score = -2.80, R) >droAna3.scaffold_12916 9428215 95 - 16180835 GGAGAAUGCCACUGCGGCGGCUGCUUUUUGUUA-----GCCAUCCAAAAAAUUCACAAAUGGUUAAAGCCAUUUAUUUCAAACAAAUCAUU--CUUCAACGA------------ ((((((((((.....)))((((((.....).))-----)))...............(((((((....)))))))..............)))--)))).....------------ ( -24.00, z-score = -2.47, R) >droWil1.scaffold_180772 5180660 99 - 8906247 ----UUUUGGAGUGCGGCA----UUUUUUGCCGC----UUUUCCCGCAGCAUUUGCAAAUGGUUAAAGUCAUUUAUUUCGAACAAAUCAUUUGCCAAAGCCACAGGAAGAG--- ----.........((((((----.....))))))----((((((.(..((.((.((((((((((....((.........))...)))))))))).)).))..).)))))).--- ( -29.70, z-score = -2.68, R) >consensus GGAGAAUGUGAAUGCGGCAGCUGCUUUUUGUUGCCACCGCCACAGGGCGCAUUCACGAAUGGUUAAAGUCAUUUAUUUCGAACAAAUCAUUUGCCACAACCCACUAACGAAGCC .((((..((((((((((((((........))))))...(((....)))))))))))((((((......)))))).))))................................... (-18.88 = -21.37 + 2.50)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:33:26 2011