| Sequence ID | dm3.chrX |
|---|---|
| Location | 17,551,341 – 17,551,404 |
| Length | 63 |
| Max. P | 0.737305 |

| Location | 17,551,341 – 17,551,404 |
|---|---|
| Length | 63 |
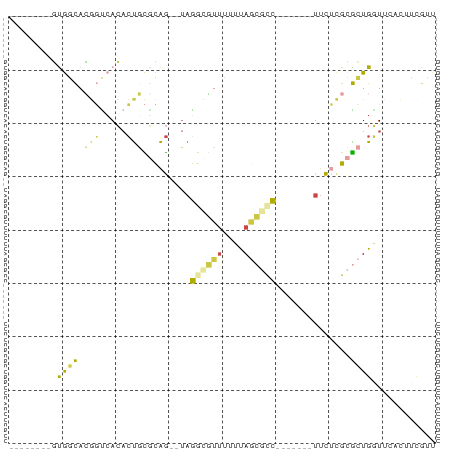
| Sequences | 6 |
| Columns | 80 |
| Reading direction | forward |
| Mean pairwise identity | 53.75 |
| Shannon entropy | 0.80387 |
| G+C content | 0.49170 |
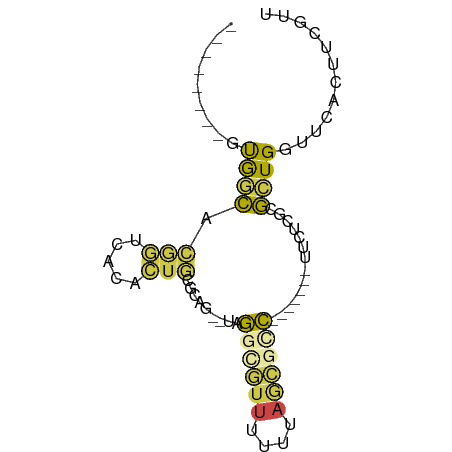
| Mean single sequence MFE | -17.75 |
| Consensus MFE | -4.94 |
| Energy contribution | -4.33 |
| Covariance contribution | -0.60 |
| Combinations/Pair | 1.92 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.28 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.737305 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 17551341 63 + 22422827 --------GUGGCACGGCCACACUGCGCAG--UAGGCGUUUUUUAGCGCC-------UUCUCGCGCUGGUUCACUUCGUU --------(((((...)))))(((((((((--.(((((((....))))))-------).)).)))).))).......... ( -26.80, z-score = -3.35, R) >droSim1.chrX 3625286 63 - 17042790 --------GUGGCACGGUCACACUGCGCAG--UAGGCGUUUUUUAGCGUC-------UUCUCGCGCUGGUUCACUUCGUU --------(((((...)))))(((((((((--.(((((((....))))))-------).)).)))).))).......... ( -21.40, z-score = -2.41, R) >droSec1.super_17 1393945 54 + 1527944 --------GUGCCACGGUCACACUGCACAG--UAGGCGUUUUUUAGCGCC-------UUCUCGCGCUGGUU--------- --------((((....).)))(((((.(((--.(((((((....))))))-------).)).).)).))).--------- ( -17.30, z-score = -1.24, R) >droYak2.chrX 16178737 63 + 21770863 --------GCGGCACGGUCACACUGCGCAG--UAGGUGUUUUUUAGCGCC-------UUCUCGCGCUGGUUCACUUCGUU --------((((...(((.((...((((((--.(((((((....))))))-------).)).))))..))..))))))). ( -20.50, z-score = -1.43, R) >droMoj3.scaffold_6473 12799655 74 - 16943266 --UCAGCUUCAUCAAAUUAAAAACCUUUGA--GCAAUUUCAAGCAAAUCUAAUGCUGUCUAUGCGAUGUU--AUUUCGAU --.......((((...(((((....)))))--(((......((((.......)))).....)))))))..--........ ( -9.30, z-score = 0.13, R) >triCas2.ChLG3 17915932 80 + 32080666 GUGUACUUUUAGUAGACUCACCCUCUGUAGAUUAAACCUUCUGCAUCACUAAAAAUUUUCAAGCACUGAUACAUUUUGUU (((((.(((((((.((.........((((((........)))))))))))))))....(((.....))))))))...... ( -11.20, z-score = -0.80, R) >consensus ________GUGGCACGGUCACACUGCGCAG__UAGGCGUUUUUUAGCGCC_______UUCUCGCGCUGGUUCACUUCGUU .........((((.(((.....))).........((((((....))))))..............))))............ ( -4.94 = -4.33 + -0.60)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:53:42 2011