| Sequence ID | dm3.chrX |
|---|---|
| Location | 17,549,799 – 17,549,919 |
| Length | 120 |
| Max. P | 0.838687 |

| Location | 17,549,799 – 17,549,919 |
|---|---|
| Length | 120 |
| Sequences | 13 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.47 |
| Shannon entropy | 0.57332 |
| G+C content | 0.53323 |
| Mean single sequence MFE | -39.57 |
| Consensus MFE | -14.52 |
| Energy contribution | -15.16 |
| Covariance contribution | 0.63 |
| Combinations/Pair | 1.93 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.838687 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 17549799 120 + 22422827 AUAUUACCCUACUCACCAGAUCCUCGGGAUGCUUGAGCGUGAGCUUAAGCAUAAACGACGGCACCUUCAGCAUAUCCACGCACAUGGACCUGUUGGCCAGCGCCUGGGAGGGAUUCAUCU ......((((.((((........(((..((((((((((....))))))))))...))).(((.(..((((((..((((......))))..))))))...).)))))))))))........ ( -45.90, z-score = -3.54, R) >droSim1.chrX 3623741 120 - 17042790 AUAUUACCCUACUCACCAGGUCCUCGGGAUGCUUGAGCGUGAGCUUGAGCAUAAACGACGGCACCUUCAGCAUAUCCACGCACAUGGACCUGUUGGCCAGCGCCUGGGAAGGAUUCAUCU .......(((..((.((((((.((((..(((((..(((....)))..)))))...))).(((.....(((((..((((......))))..)))))))).).)))))))))))........ ( -47.00, z-score = -3.42, R) >droSec1.super_17 1392442 120 + 1527944 AUAUUACCCUACUCACCAGGUCCUCGGGAUGCUUGAGCGUAAGCUUGAGCAUAAACGACGGCACCUUCAGCAUAUCCACGCACAUGGACCUGUUGGCCAGCGCCUGGGACGGAUUCAUCU .......((...((.((((((.((((..(((((..(((....)))..)))))...))).(((.....(((((..((((......))))..)))))))).).)))))))).))........ ( -44.30, z-score = -3.04, R) >droYak2.chrX 16177171 120 + 21770863 AUAUUACCCUACUCACCAGAUCCUCGGGAUGCUUGAGCGUGAGCUUGAGCAUGAACGACGGCACCUUGAGCAUGUCCACGCACAUGGACCUGUUGGCCAGCGCUUGAGAGGGAUUCAUUU ......((((.((((........(((..(((((..(((....)))..)))))...))).(((......((((.(((((......))))).)))).)))......))))))))........ ( -44.00, z-score = -2.88, R) >droEre2.scaffold_4690 7886271 120 + 18748788 AUAUUACCCUACUCACCAGAUCCUCGGGAUGCUUGAGCGUGAGCUUGAGCAUGAACGACGGCACCUUGAGCAUGUCCACGCACAUGGACCUGUUGGCCAGCGCCUGUGAGGGGUUCAUUU .....((((..(((((.......(((..(((((..(((....)))..)))))...))).(((.(..(.((((.(((((......))))).)))).)...).))).)))))))))...... ( -45.30, z-score = -2.33, R) >triCas2.ChLG6 773333 106 + 13544221 --------------AUGUCCCCCUCCCCGCCGUCCAGAGAUAGUGUGAUUGCGAGCGCGGGCAUUCGGCACAAUUCGACGCACUUGCUUCGGAUUGUCAAAGCCUGGCUCGGAUUCAUUU --------------.............(((..((....))..)))(((.(.(((((.(((((.....(.(((((((((.((....)).))))))))))...)))))))))).).)))... ( -36.50, z-score = -1.16, R) >droPer1.super_53 249878 112 - 525408 --------CCACUCACCAGAUCGUCGGAAUGCUUAAGCGCCAGCUUAAGCAUGAAGGAGGGCCCCUUCAUCAUUUCCACGCACAGCGUUCGAUUGGCCAGUGCUUGGGUGGGGUUCAUUU --------(((((((...(((((..(((((((((((((....)))))))))((((((......))))))....))))((((...)))).)))))(((....))))))))))......... ( -41.50, z-score = -2.18, R) >dp4.chrXL_group1a 1527029 112 + 9151740 --------CCACUCACCAGAUCGUCGGAAUGCUUAAGCGCCAGCUUAAGCAUGAAGGAGGGCCCCUUCAUCAUUUCCACGCACAGCGUUCGAUUGGCCAGUGCUUGGGUGGGGUUCAUUU --------(((((((...(((((..(((((((((((((....)))))))))((((((......))))))....))))((((...)))).)))))(((....))))))))))......... ( -41.50, z-score = -2.18, R) >droWil1.scaffold_180777 2115102 109 + 4753960 -----------CUUACCAAAUCCUCGGGAUGUUUGAGCGUCAGUUUAAGCAUAAACGAUGGUACCUUCAUCAUGUCAACGCACAUGGAACGAUUAGCUAAGGCUUGGGAGGGAUUCAUCU -----------..(((((.....(((..((((((((((....))))))))))...))))))))(((((.((((((......)))))).......(((....)))..)))))......... ( -30.20, z-score = -1.57, R) >droGri2.scaffold_15081 3151124 109 + 4274704 -----------CUCACCAGGUCAUCGGUAUGCUUGAGCGCUAGCUUAAGCAUAAACGAAGGCACUUUCAUCAUUUCGACGCACAUGGAGCGAUUGACCAACGCCUGCGAUGGAUUCAUCU -----------....(((((((((((((((((((((((....)))))))))))...((((....))))......))))(((.......)))..)))))..((....)).)))........ ( -33.90, z-score = -2.10, R) >droMoj3.scaffold_6328 577933 108 + 4453435 ------------UUACCAAAUCCUCGGAAUGCUUGAGUGCCAGCUUGAGCAUAAACGAUGGCACCUUCAUCAUGUCCACGCACAUGGAGCGAUUGGCCAGCGCCUGUGAGGGAUUCAUUU ------------......((((((.((((((((..(((....)))..)))))....(((((.....)))))...)))...((((.((.((.........)).)))))).))))))..... ( -32.80, z-score = -0.69, R) >droVir3.scaffold_13042 2102892 113 - 5191987 -------CUCACUCACCAGAUCGUCGGAGUGCUUGAGCGCCAGCUUGAGCAUGAACGAGGGCACCUUCAGCAUAUCGACGCACAUGGAGCGAUUGGCCAGCGCCUGGGAGGGAUUCAUUU -------.((.(((.((((((((((((.(((((..(((....)))..)))))....((((....))))......)))))((.......))))))(((....))))))))).))....... ( -39.70, z-score = -0.84, R) >anoGam1.chr2R 31452301 106 - 62725911 --------------ACCAGAUUCUGGGGAUCCUGCAGACUCAGCUUGAUCAUAAGCGAGGGCAUCUUUACCGUUUCCACGCAGAUCGUGCGCAUCGACAGCGCUUGGCUGGGGUUCAUCU --------------.((((...))))(((((((((...(((.(((((....)))))))).))).................(((.(((.((((.......)))).)))))))))))).... ( -31.80, z-score = 0.19, R) >consensus ________C_ACUCACCAGAUCCUCGGGAUGCUUGAGCGUCAGCUUGAGCAUAAACGACGGCACCUUCAGCAUUUCCACGCACAUGGACCGAUUGGCCAGCGCCUGGGAGGGAUUCAUUU ...............((...........((((((((((....)))))))))).......(((.................................)))............))........ (-14.52 = -15.16 + 0.63)

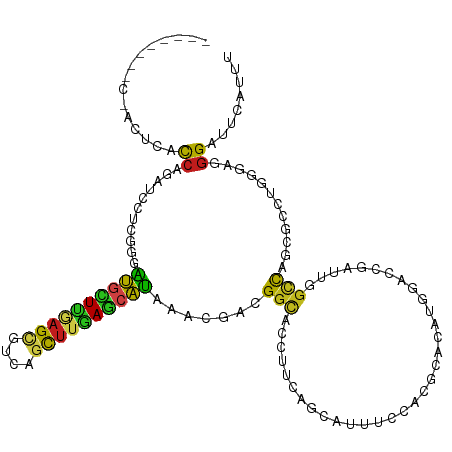

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:53:41 2011