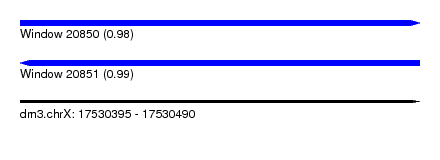
| Sequence ID | dm3.chrX |
|---|---|
| Location | 17,530,395 – 17,530,490 |
| Length | 95 |
| Max. P | 0.993036 |

| Location | 17,530,395 – 17,530,490 |
|---|---|
| Length | 95 |
| Sequences | 7 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 67.80 |
| Shannon entropy | 0.64317 |
| G+C content | 0.59804 |
| Mean single sequence MFE | -37.43 |
| Consensus MFE | -20.08 |
| Energy contribution | -21.80 |
| Covariance contribution | 1.72 |
| Combinations/Pair | 1.47 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.95 |
| SVM RNA-class probability | 0.976278 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
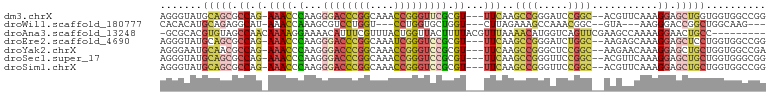
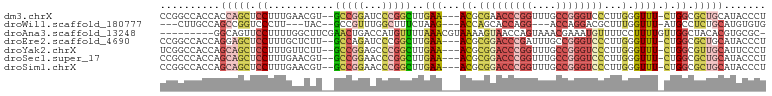
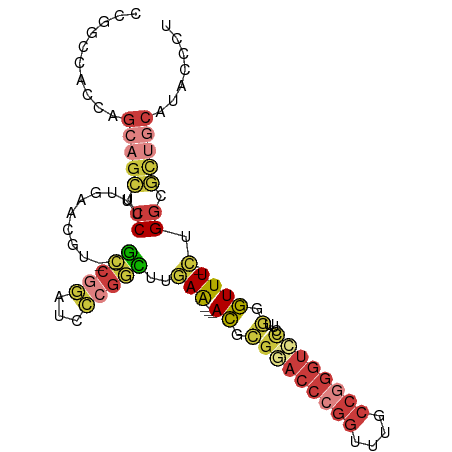
>dm3.chrX 17530395 95 + 22422827 AGGGUAUGCAGCGCCAG-AAACCCAAGGGACCCGGCAAACCGGGUUCGCGU---UUCAAGCCGGGAUCCGGC--ACGUUCAAAGGAGCUGGUGGUGGCCGG ..(((...((.((((((-....((...((((((((....))))))))((((---.....(((((...)))))--)))).....))..)))))).))))).. ( -40.70, z-score = -1.14, R) >droWil1.scaffold_180777 2094992 86 + 4753960 CACACAUGCAGAGGCAU-AAACCAAAGCGUCCUGGU---CCUGGUGCUGGU---CUUAGAAAGCCAAACGGC--GUA---AAGGGACCGGCUGGCAAG--- ......(((........-..((((..((......))---..))))((((((---(((.....(((....)))--...---..)))))))))..)))..--- ( -26.90, z-score = -0.42, R) >droAna3.scaffold_13248 3017299 91 - 4840945 -GCGCACGUGUAGCCAACAAAAGGAAAACAUUUCGUUUACUGGUUACUUUUACGUUUAAAACAUGGUCAGUUCGAAGCCAAAAGGAACUGCC--------- -(((.....(((((((...(((.(((.....))).)))..))))))).....))).........((.((((((...........))))))))--------- ( -19.40, z-score = -0.43, R) >droEre2.scaffold_4690 7866945 95 + 18748788 AGGGUAUGCAGCGCCAG-AAACCCAAGGGACCCGGCAAAUCGGGUCCGCGU---UUCAAGCCGGGAUCUGGC--AAGAGCAAAGGAGCUCCUGGUGGCCGG .((.(((.(((.(((((-(..(((...((((((((....))))))))((..---.....)).))).))))))--..((((......)))))))))).)).. ( -43.10, z-score = -2.12, R) >droYak2.chrX 16157491 95 + 21770863 AGGGAAUGCAACGCCAG-AAACCCAAGGGACCCGGCAAACCGGGUCCGCGU---UUCAAGCCGGGCUCCGGC--AAGAACAAAGGAGCUGCUGGUGGCCGA ..((..((.(((((..(-.....)...((((((((....))))))))))))---).))..)).(((((((((--(.(..(....)..))))))).)))).. ( -40.40, z-score = -1.23, R) >droSec1.super_17 1373223 95 + 1527944 AGGGUAUGCAGCGCCAG-AAACCCAAGGGACCCGGCAAACCGGGUCCGCGU---UUCAAGCCGGGUUCCGGC--ACGUUCAAAGGAGCUGCUGGUGGGCGG ...........((((..-.(((((...((((((((....))))))))((..---.....)).)))))(((((--(.((((....))))))))))..)))). ( -45.50, z-score = -2.11, R) >droSim1.chrX 3603148 95 - 17042790 AGGGUAUGCAGCGCCAG-AAACCCAAGGGACCCGGCAAACCGGGUCCGCGU---UUCAAGCCGGGUUCCGGC--ACGUUCAAAGGAGCUGCUGGUGGCCGG .((.(((.(((((((.(-((.(((...((((((((....))))))))((..---.....)).)))))).)))--..((((....)))).))))))).)).. ( -46.00, z-score = -2.26, R) >consensus AGGGUAUGCAGCGCCAG_AAACCCAAGGGACCCGGCAAACCGGGUCCGCGU___UUCAAGCCGGGAUCCGGC__AAGUUCAAAGGAGCUGCUGGUGGCCGG .......(((((.((............((((((((....))))))))............((((.....))))...........)).))))).......... (-20.08 = -21.80 + 1.72)
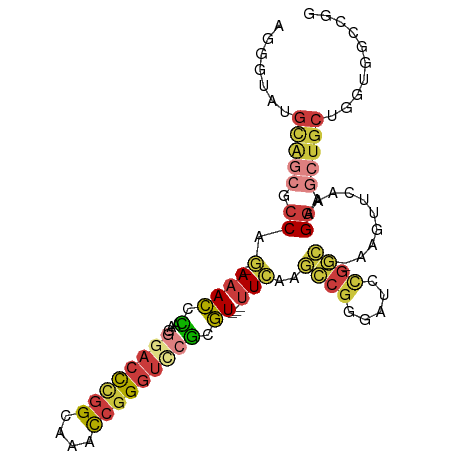
| Location | 17,530,395 – 17,530,490 |
|---|---|
| Length | 95 |
| Sequences | 7 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 67.80 |
| Shannon entropy | 0.64317 |
| G+C content | 0.59804 |
| Mean single sequence MFE | -35.73 |
| Consensus MFE | -19.93 |
| Energy contribution | -21.91 |
| Covariance contribution | 1.99 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.58 |
| SVM RNA-class probability | 0.993036 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 17530395 95 - 22422827 CCGGCCACCACCAGCUCCUUUGAACGU--GCCGGAUCCCGGCUUGAA---ACGCGAACCCGGUUUGCCGGGUCCCUUGGGUUU-CUGGCGCUGCAUACCCU ..((.......((((.((.........--(((((...)))))..(((---((.(((((((((....))))))...))).))))-).)).)))).....)). ( -35.40, z-score = -0.86, R) >droWil1.scaffold_180777 2094992 86 - 4753960 ---CUUGCCAGCCGGUCCCUU---UAC--GCCGUUUGGCUUUCUAAG---ACCAGCACCAGG---ACCAGGACGCUUUGGUUU-AUGCCUCUGCAUGUGUG ---...(((((.((((.....---...--)))).)))))........---....((((..((---((((((....))))))))-((((....)))))))). ( -23.80, z-score = -0.10, R) >droAna3.scaffold_13248 3017299 91 + 4840945 ---------GGCAGUUCCUUUUGGCUUCGAACUGACCAUGUUUUAAACGUAAAAGUAACCAGUAAACGAAAUGUUUUCCUUUUGUUGGCUACACGUGCGC- ---------((((((((...........)))))).))..........((((...(((.((((((((.(((.....)))..)))))))).)))...)))).- ( -18.70, z-score = -0.00, R) >droEre2.scaffold_4690 7866945 95 - 18748788 CCGGCCACCAGGAGCUCCUUUGCUCUU--GCCAGAUCCCGGCUUGAA---ACGCGGACCCGAUUUGCCGGGUCCCUUGGGUUU-CUGGCGCUGCAUACCCU .((((....((((((......))))))--((((((.((((((.....---..))(((((((......)))))))..))))..)-)))))))))........ ( -43.40, z-score = -3.39, R) >droYak2.chrX 16157491 95 - 21770863 UCGGCCACCAGCAGCUCCUUUGUUCUU--GCCGGAGCCCGGCUUGAA---ACGCGGACCCGGUUUGCCGGGUCCCUUGGGUUU-CUGGCGUUGCAUUCCCU ..........(((((.((.........--(((((...)))))..(((---((.(((((((((....))))))))...).))))-).)).)))))....... ( -41.20, z-score = -1.81, R) >droSec1.super_17 1373223 95 - 1527944 CCGCCCACCAGCAGCUCCUUUGAACGU--GCCGGAACCCGGCUUGAA---ACGCGGACCCGGUUUGCCGGGUCCCUUGGGUUU-CUGGCGCUGCAUACCCU ..........(((((((....))....--(((((((((((((.....---..))((((((((....))))))))..)))).))-))))))))))....... ( -43.80, z-score = -2.97, R) >droSim1.chrX 3603148 95 + 17042790 CCGGCCACCAGCAGCUCCUUUGAACGU--GCCGGAACCCGGCUUGAA---ACGCGGACCCGGUUUGCCGGGUCCCUUGGGUUU-CUGGCGCUGCAUACCCU ..........(((((((....))....--(((((((((((((.....---..))((((((((....))))))))..)))).))-))))))))))....... ( -43.80, z-score = -2.53, R) >consensus CCGGCCACCAGCAGCUCCUUUGAACGU__GCCGGAUCCCGGCUUGAA___ACGCGGACCCGGUUUGCCGGGUCCCUUGGGUUU_CUGGCGCUGCAUACCCU ..........(((((.((...........(((((...)))))............((((((((....))))))))............)).)))))....... (-19.93 = -21.91 + 1.99)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:53:36 2011