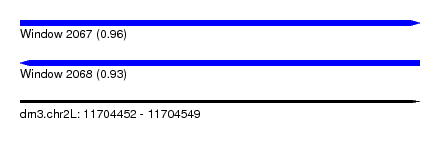
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 11,704,452 – 11,704,549 |
| Length | 97 |
| Max. P | 0.961878 |

| Location | 11,704,452 – 11,704,549 |
|---|---|
| Length | 97 |
| Sequences | 9 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 84.32 |
| Shannon entropy | 0.30593 |
| G+C content | 0.41642 |
| Mean single sequence MFE | -29.99 |
| Consensus MFE | -21.49 |
| Energy contribution | -21.20 |
| Covariance contribution | -0.29 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.70 |
| SVM RNA-class probability | 0.961878 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
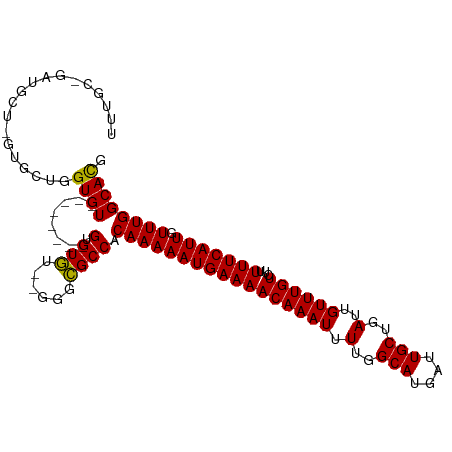
>dm3.chr2L 11704452 97 + 23011544 UUUGCUGAUGCUCGUGCUGGUGU--------GUGUG----GGCGCCACAAAAAUGAAAACAAAUUUUGGCAUGAUUGCUGAUUGUUUGUCUUUUUCAUUGUUUGGCACG .........(((((..(......--------)..))----)))((((....(((((((((((((.(..(((....)))..)..))))))...)))))))...))))... ( -31.40, z-score = -2.72, R) >droSim1.chr2L 11513906 101 + 22036055 UUUGCCGAUGCUCGUGCUGGUGU--------GUGUGUGUGGGCGCCACAAAAAUGAAAACAAAUUUUGGCAUGAUUGCUGAUUGUUUGUCUUUUUCAUUGUUUGGCACG ...(((.((((.((..(....).--------.)).)))).)))((((....(((((((((((((.(..(((....)))..)..))))))...)))))))...))))... ( -35.80, z-score = -3.32, R) >droSec1.super_3 7102002 99 + 7220098 UUUGCCGAUGCUCGUGCUGGUGU--------GUGUGU--GGGCGCCACAAAAAUGAAAACAAAUUUUGGCAUGAUUGCUGAUUGUUUGUCUUUUUCAUUGUUUGGCACG ..((((((.(((((..(......--------..)..)--))))........(((((((((((((.(..(((....)))..)..))))))...)))))))..)))))).. ( -31.30, z-score = -2.03, R) >droYak2.chr2L 8128626 109 + 22324452 UUUGCUGAUGCUGGUGCUGGUGUUAGUGCUGGUGUGUGUGGGCGCCACAAAAAUGAAAACAAAUUUUGGCAUGAUUGCUGAUUGUUUGUCUUUUUCAUUGUUUGGCACG ...(((.((((..(..(((....)))..)..))....)).)))((((....(((((((((((((.(..(((....)))..)..))))))...)))))))...))))... ( -34.70, z-score = -2.37, R) >droEre2.scaffold_4929 12935319 94 - 26641161 UUUGCCGAUG---GUGCUGGUGU--------GUGUG----GGCGCCACAAAAAUGAAAACAAAUUUUGGCAUGAUUGCUGAUUGUUUGUCUUUUUCAUUGUUUGGCACG ..((((((((---(((((.(...--------...).----)))))))....(((((((((((((.(..(((....)))..)..))))))...)))))))..)))))).. ( -31.80, z-score = -2.77, R) >droAna3.scaffold_12916 9421030 84 - 16180835 ---------------GUGGGUGU--------GUGUAU--GGGUGCCACAAAAAUGAAAACAAAUUUUGGCAUGAUUGCUGAUUGUUUGUCUUUUUCAUUGUUUGGCACG ---------------........--------......--..((((((....(((((((((((((.(..(((....)))..)..))))))...)))))))...)))))). ( -25.20, z-score = -2.36, R) >dp4.chr4_group3 2570936 93 + 11692001 ------UGUGUGUUUGG--GUGUGU------GUGUGU--GGGCGCCACAAAAAUGAAAACAAAUUUUGGCAUGAUUGCUGAUUGUUUGUCUUUUUCAUUGUUUGGCACG ------.((((.....(--((((.(------......--).))))).(((((((((((((((((.(..(((....)))..)..))))))...))))))).)))))))). ( -27.80, z-score = -2.04, R) >droPer1.super_1 4049604 95 + 10282868 ------UGUGUGUUUGGCUGUGUGU------GUGUGU--GGGCGCCACAAAAAUGAAAACAAAUUUUGGCAUGAUUGCUGAUUGUUUGUCUUUUUCAUUGUUUGGCACG ------...(((((..((((((.((------((....--..))))))))..(((((((((((((.(..(((....)))..)..))))))...)))))))))..))))). ( -28.70, z-score = -2.06, R) >droWil1.scaffold_180772 3746066 88 + 8906247 --------------UGUGUGUGUGU------GAGUGUA-GGGCGCCACAAAAAUGAAAACAAAUUUUGGCAUGAUUGCUGAUUGUUUGUCUUUUUCAUUGUUUGGCAUG --------------...((((.(((------(.(((..-...)))))))..(((((((((((((.(..(((....)))..)..))))))...))))))).....)))). ( -23.20, z-score = -1.23, R) >consensus UUUGC_GAUGCU_GUGCUGGUGU________GUGUGU__GGGCGCCACAAAAAUGAAAACAAAUUUUGGCAUGAUUGCUGAUUGUUUGUCUUUUUCAUUGUUUGGCACG ...................((((........(.(((......)))).(((((((((((((((((.(..(((....)))..)..))))))...))))))).)))))))). (-21.49 = -21.20 + -0.29)

| Location | 11,704,452 – 11,704,549 |
|---|---|
| Length | 97 |
| Sequences | 9 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 84.32 |
| Shannon entropy | 0.30593 |
| G+C content | 0.41642 |
| Mean single sequence MFE | -18.79 |
| Consensus MFE | -17.38 |
| Energy contribution | -17.39 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.92 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.37 |
| SVM RNA-class probability | 0.931825 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
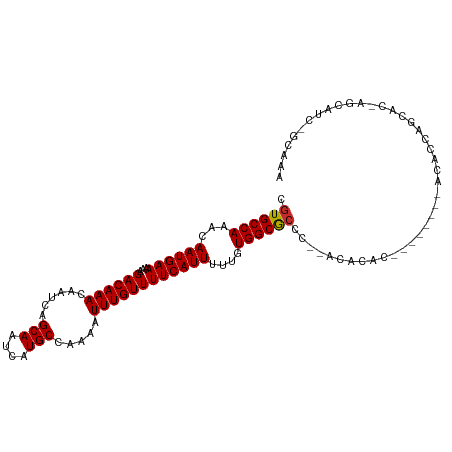
>dm3.chr2L 11704452 97 - 23011544 CGUGCCAAACAAUGAAAAAGACAAACAAUCAGCAAUCAUGCCAAAAUUUGUUUUCAUUUUUGUGGCGCC----CACAC--------ACACCAGCACGAGCAUCAGCAAA (((((.....((((((...((((((......(((....))).....))))))))))))..(((((...)----)))).--------......))))).((....))... ( -19.90, z-score = -1.31, R) >droSim1.chr2L 11513906 101 - 22036055 CGUGCCAAACAAUGAAAAAGACAAACAAUCAGCAAUCAUGCCAAAAUUUGUUUUCAUUUUUGUGGCGCCCACACACAC--------ACACCAGCACGAGCAUCGGCAAA .((((((...((((((...((((((......(((....))).....))))))))))))....))))))..........--------......((.((.....))))... ( -21.50, z-score = -1.22, R) >droSec1.super_3 7102002 99 - 7220098 CGUGCCAAACAAUGAAAAAGACAAACAAUCAGCAAUCAUGCCAAAAUUUGUUUUCAUUUUUGUGGCGCCC--ACACAC--------ACACCAGCACGAGCAUCGGCAAA .((((((...((((((...((((((......(((....))).....))))))))))))....))))))..--......--------......((.((.....))))... ( -21.50, z-score = -1.23, R) >droYak2.chr2L 8128626 109 - 22324452 CGUGCCAAACAAUGAAAAAGACAAACAAUCAGCAAUCAUGCCAAAAUUUGUUUUCAUUUUUGUGGCGCCCACACACACCAGCACUAACACCAGCACCAGCAUCAGCAAA .((((((...((((((...((((((......(((....))).....))))))))))))....))))))............((..........))....((....))... ( -19.30, z-score = -0.89, R) >droEre2.scaffold_4929 12935319 94 + 26641161 CGUGCCAAACAAUGAAAAAGACAAACAAUCAGCAAUCAUGCCAAAAUUUGUUUUCAUUUUUGUGGCGCC----CACAC--------ACACCAGCAC---CAUCGGCAAA .((((((...((((((...((((((......(((....))).....))))))))))))....)))))).----.....--------......((..---.....))... ( -18.60, z-score = -0.85, R) >droAna3.scaffold_12916 9421030 84 + 16180835 CGUGCCAAACAAUGAAAAAGACAAACAAUCAGCAAUCAUGCCAAAAUUUGUUUUCAUUUUUGUGGCACCC--AUACAC--------ACACCCAC--------------- .((((((...((((((...((((((......(((....))).....))))))))))))....))))))..--......--------........--------------- ( -18.20, z-score = -2.72, R) >dp4.chr4_group3 2570936 93 - 11692001 CGUGCCAAACAAUGAAAAAGACAAACAAUCAGCAAUCAUGCCAAAAUUUGUUUUCAUUUUUGUGGCGCCC--ACACAC------ACACAC--CCAAACACACA------ .((((((...((((((...((((((......(((....))).....))))))))))))....))))))..--......------......--...........------ ( -17.80, z-score = -1.95, R) >droPer1.super_1 4049604 95 - 10282868 CGUGCCAAACAAUGAAAAAGACAAACAAUCAGCAAUCAUGCCAAAAUUUGUUUUCAUUUUUGUGGCGCCC--ACACAC------ACACACAGCCAAACACACA------ .((((((...((((((...((((((......(((....))).....))))))))))))....))))))..--......------...................------ ( -17.80, z-score = -1.47, R) >droWil1.scaffold_180772 3746066 88 - 8906247 CAUGCCAAACAAUGAAAAAGACAAACAAUCAGCAAUCAUGCCAAAAUUUGUUUUCAUUUUUGUGGCGCCC-UACACUC------ACACACACACA-------------- ...((((...((((((...((((((......(((....))).....))))))))))))....))))....-.......------...........-------------- ( -14.50, z-score = -1.25, R) >consensus CGUGCCAAACAAUGAAAAAGACAAACAAUCAGCAAUCAUGCCAAAAUUUGUUUUCAUUUUUGUGGCGCCC__ACACAC________ACACCAGCAC_AGCAUC_GCAAA .((((((...((((((...((((((......(((....))).....))))))))))))....))))))......................................... (-17.38 = -17.39 + 0.01)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:33:24 2011