| Sequence ID | dm3.chrX |
|---|---|
| Location | 17,505,078 – 17,505,232 |
| Length | 154 |
| Max. P | 0.879682 |

| Location | 17,505,078 – 17,505,192 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 94.43 |
| Shannon entropy | 0.09765 |
| G+C content | 0.41267 |
| Mean single sequence MFE | -32.35 |
| Consensus MFE | -22.06 |
| Energy contribution | -23.14 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.85 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.803204 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
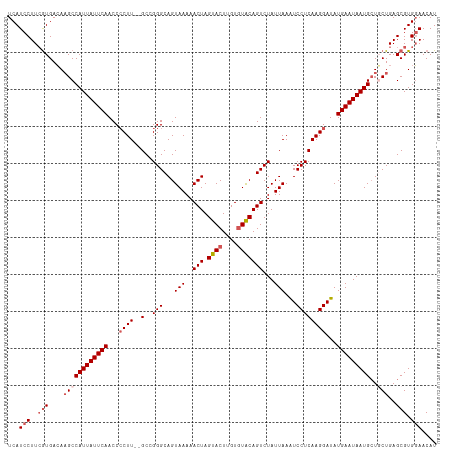
>dm3.chrX 17505078 114 + 22422827 UCAUCCUUCGUGACAAGCCAUUAUUCAACCCCUU--GCCGGGCAGUAAAAACUAGUACUUGUGUACAGUCUAUUAAAUCCUCAAGGAUAUGAAUAAUGCUGCUGAGCGUGGAACAU ...(((..(((....((((((((((((...((((--(..(((...(((..(((.((((....)))))))...)))...))))))))...)))))))))..)))..))).))).... ( -31.80, z-score = -2.77, R) >droYak2.chrX 16132696 116 + 21770863 UCAUCCUUCAUGAGAAGCCAUUAUUCAACCCCUUUUGCAGGGCAGUAAAAACUAGUACUAGUUUGCAGUCUAUUAAAUCCUCAAGGAUAUGAAUAAUGCUUCUGAGCGUGCAACAU ........((((((((((.((((((((.......(((((((..((((........))))..)))))))........((((....)))).))))))))))))))...))))...... ( -27.90, z-score = -1.92, R) >droEre2.scaffold_4690 7841928 114 + 18748788 UCAUCCUUAGUGACACGCCAUUAUUCAACCCCUU--GCAGGGCAGUAAAAACUAGUACUUGUGUACAGUCUAUUAAGUCCUCAAGGAUAUGAAUAAUGCUUCUGAGCGUGGAACAU .........((..((((((((((((((...((((--(.(((((.......(((.((((....))))))).......))))))))))...))))))))).......)))))..)).. ( -37.25, z-score = -4.23, R) >droSec1.super_17 1347884 114 + 1527944 UCAUCCUUCGUGACAAGCCAUUAUUCAACCGCUU--GCCGGGCAGUAAAAACUAGUACUUGUGUACAGUCUAUUAAAUCCUCAAGGAUAUGAAUAAUGGUGCUGAGCGUGGAACAU ...(((..(((.....(((((((((((.......--...(((((((....))).((((....)))).)))).....((((....)))).))))))))))).....))).))).... ( -33.00, z-score = -2.57, R) >droSim1.chrX_random 4493276 114 + 5698898 UCAUCCUUCGUGACAAGCCAUUAUUCAACCCCUU--GCCGGGCAGUAAAAACUAGUACUUGUGUACAGUCUAUUAAAUCCUCAAGGAUAUGAAUAAUGCUGCUGAGCGUGGAACAU ...(((..(((....((((((((((((...((((--(..(((...(((..(((.((((....)))))))...)))...))))))))...)))))))))..)))..))).))).... ( -31.80, z-score = -2.77, R) >consensus UCAUCCUUCGUGACAAGCCAUUAUUCAACCCCUU__GCCGGGCAGUAAAAACUAGUACUUGUGUACAGUCUAUUAAAUCCUCAAGGAUAUGAAUAAUGCUGCUGAGCGUGGAACAU ...(((..(((....((((((((((((............(((((((....))).((((....)))).)))).....((((....)))).)))))))))..)))..))).))).... (-22.06 = -23.14 + 1.08)


| Location | 17,505,112 – 17,505,232 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 72.06 |
| Shannon entropy | 0.55906 |
| G+C content | 0.41873 |
| Mean single sequence MFE | -30.51 |
| Consensus MFE | -16.50 |
| Energy contribution | -17.53 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.879682 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
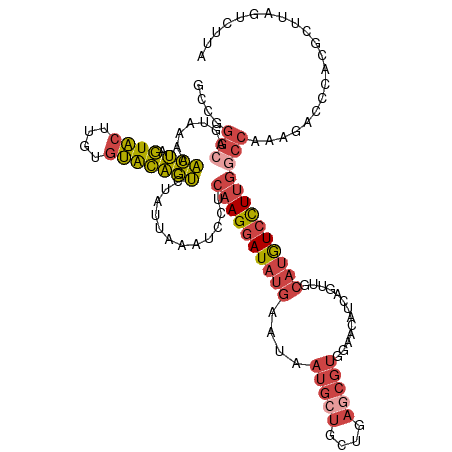
>dm3.chrX 17505112 120 + 22422827 GCCGGGCAGUAAAAACUAGUACUUGUGUACAGUCUAUUAAAUCCUCAAGGAUAUGAAUAAUGCUGCUGAGCGUGGAACAUCAGUUGCAUGUCCUUGGCCAAAGACCCACGCUUAGUCUUA (((((((((((...(((.((((....)))))))...(((.((((....)))).)))....)))))))(..((((.(((....))).))))..).))))..(((((.........))))). ( -34.10, z-score = -1.56, R) >droAna3.scaffold_13248 1927816 101 - 4840945 -------------ACAGGAUUCUUAUGGAUCUGAUAUAAAGGAUUCAAGGAUAAAAAUA------CUAAGUGUGAGCCUACUUUUCAAGAUAUUUGAGCCUGGAGUUCCUUUGAAAUUUC -------------...((((((....))))))....((((((((((.(((....(((((------(((((((......)))))....)).)))))...))).))).)))))))....... ( -21.40, z-score = -0.48, R) >droYak2.chrX 16132732 120 + 21770863 GCAGGGCAGUAAAAACUAGUACUAGUUUGCAGUCUAUUAAAUCCUCAAGGAUAUGAAUAAUGCUUCUGAGCGUGCAACAUCAGUUGCAUGUCCUCAGCCAAAGACCCACGGUAAGUGUUA (((((..((((........))))..))))).((((.(((.((((....)))).))).........(((((((((((((....))))))))..)))))....)))).(((.....)))... ( -31.80, z-score = -1.46, R) >droEre2.scaffold_4690 7841962 101 + 18748788 GCAGGGCAGUAAAAACUAGUACUUGUGUACAGUCUAUUAAGUCCUCAAGGAUAUGAAUAAUGCUUCUGAGCGUGGAACAUCAGUGGCAUGUCCUUGUCCAA------------------- (.(((((.......(((.((((....))))))).......))))))..(((((.((...(((((.((((..(.....).)))).))))).))..)))))..------------------- ( -28.34, z-score = -1.59, R) >droSec1.super_17 1347918 120 + 1527944 GCCGGGCAGUAAAAACUAGUACUUGUGUACAGUCUAUUAAAUCCUCAAGGAUAUGAAUAAUGGUGCUGAGCGUGGAACAUCAGUUGCAUGUCCUUGGCCAAAGACCCACGCUUAGUCUUA ((((((((((....))).((((....)))).)))).(((.((((....)))).))).....)))((((((((((((((....)))....(((.((....)).)))))))))))))).... ( -34.20, z-score = -1.43, R) >droSim1.chrX_random 4493310 120 + 5698898 GCCGGGCAGUAAAAACUAGUACUUGUGUACAGUCUAUUAAAUCCUCAAGGAUAUGAAUAAUGCUGCUGAGCGUGGAACAUCAGUUGCAUGUCCUUGGCCAAAGACCCACGCUUAGUCUAA (((((((((((...(((.((((....)))))))...(((.((((....)))).)))....)))))))(..((((.(((....))).))))..).))))...((((.........)))).. ( -33.20, z-score = -1.36, R) >consensus GCCGGGCAGUAAAAACUAGUACUUGUGUACAGUCUAUUAAAUCCUCAAGGAUAUGAAUAAUGCUGCUGAGCGUGGAACAUCAGUUGCAUGUCCUUGGCCAAAGACCCACGCUUAGUCUUA ....(((.......(((.((((....)))))))............((((((((((....(((((....))))).............)))))))))))))..................... (-16.50 = -17.53 + 1.03)

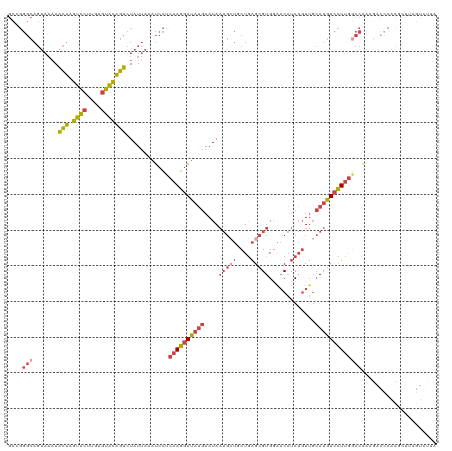
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:53:29 2011