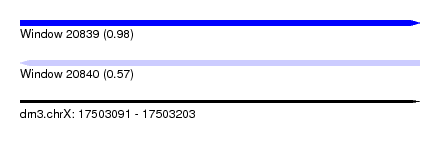
| Sequence ID | dm3.chrX |
|---|---|
| Location | 17,503,091 – 17,503,203 |
| Length | 112 |
| Max. P | 0.981319 |

| Location | 17,503,091 – 17,503,203 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 72.46 |
| Shannon entropy | 0.51263 |
| G+C content | 0.48000 |
| Mean single sequence MFE | -35.30 |
| Consensus MFE | -21.19 |
| Energy contribution | -22.86 |
| Covariance contribution | 1.67 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.07 |
| SVM RNA-class probability | 0.981319 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
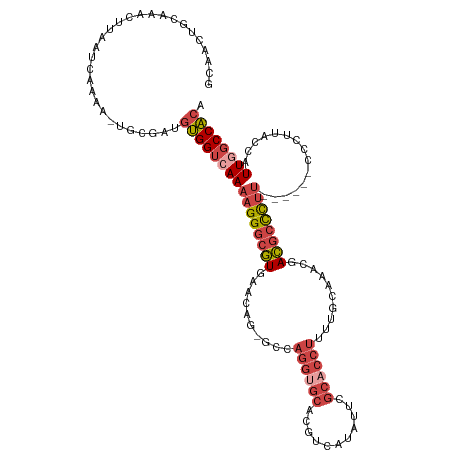
>dm3.chrX 17503091 112 + 22422827 GCAACUGCAAAGUUAAUCAAAA-UGCAAUGUGGUCAAAAGGGCGUGAACAG-GCCAGGUGCACGUCAUAUUCGCACCUUUUGUAAACGACGCCCUU------CCCUUAACAUUCGCCACA ..((((....))))........-.(((((((......(((((((((.((((-(..((((((...........)))))))))))...).))))))))------......))))).)).... ( -29.40, z-score = -1.06, R) >droSim1.chrX_random 4490667 112 + 5698898 GCAACUGCAAAGUUAAUCAAAA-UGCGAUGUGGUCAAAAGGGCGUGAACAG-GCCAGGUGCACGUCAUAUUCGCACCUUUUGCGAACGACGCCCUU------UCCUUACCGUUGGCCACA ..((((....))))........-.....((((((((((((((((((.((..-.....)).)))(((...((((((.....)))))).)))))))))------.........))))))))) ( -34.50, z-score = -1.05, R) >droSec1.super_17 1345997 111 + 1527944 GCAACUGCGAGGU-AAUCGAA--UGCGAUGUGGUCAAAAGGGCGUGACCGUUGGCAGGUGCACGUCAUAUUCGCACCUUUUGCGAACGACGCCCUU------UCCUUACCGUUGGCCACA (((....(((...-..)))..--)))..(((((((((((((((((...((((.((((((((...........)))))...))).))))))))))))------.........))))))))) ( -43.70, z-score = -2.42, R) >droYak2.chrX 16130842 118 + 21770863 UCAACUGGGAACUUAAUCAAAA-UGCGAUGUGGUCAAAAGGGCGUUAACAG-GCCAGGUGCAAGUCAUAUUUGCACCUUUUGCAAACGACGCCCUUGCGACGCUUUUUGCAUUGGCCACA ......................-.....((((((((((((((((((.....-((.((((((((((...))))))))))...))....)))))))))((((......)))).))))))))) ( -43.90, z-score = -3.43, R) >droEre2.scaffold_4690 7840278 115 + 18748788 ---ACUGCAAACUUAAUCAAAA-UACGUUGUGGUCAAAAGGGCGUUAACAG-GCCAGGCGCAAGUCAUAUUUGCACCUUUUGCAUAUGACGCCCUUACGACGCUUUUUGCAUUGGCCACA ---...................-.....(((((((((((((((((((....-((.(((.((((((...)))))).)))...))...)))))))))).....((.....)).))))))))) ( -40.30, z-score = -3.64, R) >droAna3.scaffold_13248 1920133 102 - 4840945 GGUGCUGCAAGCUUAAUCAAAAAUUCUCUGCGGUCAAGUGGCCAUUUCCAG-----------UGUCCUGU-CACACCUUUUGGAUUGGAUUUUUUGG----AUUUCUUUCAUUUUCCG-- ((.((((((...................)))))).((((((.....(((((-----------.((((.((-(.((.....))))).))))...))))----)......)))))).)).-- ( -20.01, z-score = 0.30, R) >consensus GCAACUGCAAACUUAAUCAAAA_UGCGAUGUGGUCAAAAGGGCGUGAACAG_GCCAGGUGCACGUCAUAUUCGCACCUUUUGCAAACGACGCCCUU______CCCUUACCAUUGGCCACA .............................((((((((((((((((..........((((((...........))))))..........))))))))...............)))))))). (-21.19 = -22.86 + 1.67)

| Location | 17,503,091 – 17,503,203 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 72.46 |
| Shannon entropy | 0.51263 |
| G+C content | 0.48000 |
| Mean single sequence MFE | -33.20 |
| Consensus MFE | -19.57 |
| Energy contribution | -21.58 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.17 |
| Mean z-score | -0.91 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.570724 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
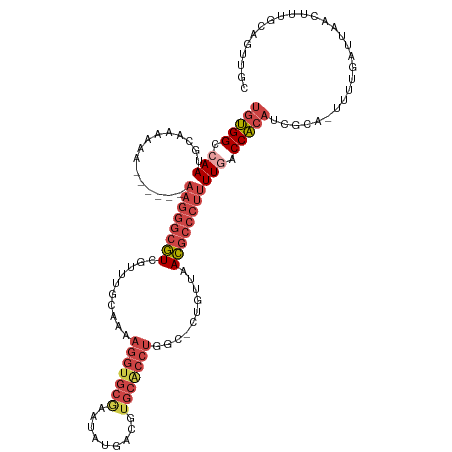
>dm3.chrX 17503091 112 - 22422827 UGUGGCGAAUGUUAAGGG------AAGGGCGUCGUUUACAAAAGGUGCGAAUAUGACGUGCACCUGGC-CUGUUCACGCCCUUUUGACCACAUUGCA-UUUUGAUUAACUUUGCAGUUGC ....(((((.((((((((------((((((((.(...(((..(((((((.........)))))))...-.))).))))))))))...)).((.....-...)).))))))))))...... ( -33.60, z-score = -0.89, R) >droSim1.chrX_random 4490667 112 - 5698898 UGUGGCCAACGGUAAGGA------AAGGGCGUCGUUCGCAAAAGGUGCGAAUAUGACGUGCACCUGGC-CUGUUCACGCCCUUUUGACCACAUCGCA-UUUUGAUUAACUUUGCAGUUGC ......(((((((...((------((((((((.....((...(((((((.........))))))).))-......)))))))))).))).....(((-.............))).)))). ( -36.12, z-score = -0.89, R) >droSec1.super_17 1345997 111 - 1527944 UGUGGCCAACGGUAAGGA------AAGGGCGUCGUUCGCAAAAGGUGCGAAUAUGACGUGCACCUGCCAACGGUCACGCCCUUUUGACCACAUCGCA--UUCGAUU-ACCUCGCAGUUGC ......((((.(..((((------((((((((((((((((.....)))))..)))))(((.(((.......))))))))))))).......((((..--..)))).-.)))..).)))). ( -37.90, z-score = -1.02, R) >droYak2.chrX 16130842 118 - 21770863 UGUGGCCAAUGCAAAAAGCGUCGCAAGGGCGUCGUUUGCAAAAGGUGCAAAUAUGACUUGCACCUGGC-CUGUUAACGCCCUUUUGACCACAUCGCA-UUUUGAUUAAGUUCCCAGUUGA ((((......((.....))((((.((((((((.....((...((((((((.......)))))))).))-......)))))))).)))))))).....-...................... ( -36.60, z-score = -1.27, R) >droEre2.scaffold_4690 7840278 115 - 18748788 UGUGGCCAAUGCAAAAAGCGUCGUAAGGGCGUCAUAUGCAAAAGGUGCAAAUAUGACUUGCGCCUGGC-CUGUUAACGCCCUUUUGACCACAACGUA-UUUUGAUUAAGUUUGCAGU--- ((((......((.....))((((.((((((((.(((.((...((((((((.......)))))))).))-.)))..)))))))).))))))))..(((-..((.....))..)))...--- ( -38.80, z-score = -2.30, R) >droAna3.scaffold_13248 1920133 102 + 4840945 --CGGAAAAUGAAAGAAAU----CCAAAAAAUCCAAUCCAAAAGGUGUG-ACAGGACA-----------CUGGAAAUGGCCACUUGACCGCAGAGAAUUUUUGAUUAAGCUUGCAGCACC --.(((...(....)...)----))..................(((((.-.((((...-----------.(((......)))(((((...(((((...))))).)))))))))..))))) ( -16.20, z-score = 0.96, R) >consensus UGUGGCCAAUGCAAAAAA______AAGGGCGUCGUUUGCAAAAGGUGCGAAUAUGACGUGCACCUGGC_CUGUUAACGCCCUUUUGACCACAUCGCA_UUUUGAUUAACUUUGCAGUUGC (((((.(((...............((((((((..........(((((((.........)))))))..........))))))))))).)))))............................ (-19.57 = -21.58 + 2.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:53:27 2011