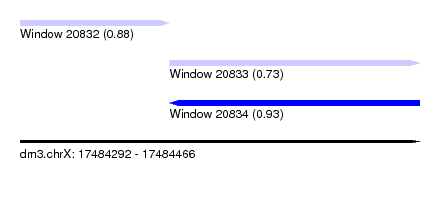
| Sequence ID | dm3.chrX |
|---|---|
| Location | 17,484,292 – 17,484,466 |
| Length | 174 |
| Max. P | 0.928684 |

| Location | 17,484,292 – 17,484,357 |
|---|---|
| Length | 65 |
| Sequences | 6 |
| Columns | 65 |
| Reading direction | forward |
| Mean pairwise identity | 79.18 |
| Shannon entropy | 0.39229 |
| G+C content | 0.43590 |
| Mean single sequence MFE | -12.29 |
| Consensus MFE | -6.62 |
| Energy contribution | -8.53 |
| Covariance contribution | 1.92 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.883298 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
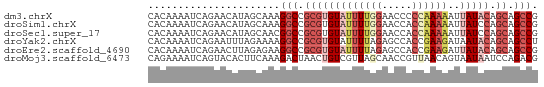
>dm3.chrX 17484292 65 + 22422827 CACAAAAUCAGAACAUAGCAAAGGCCGCGUGUAUUUUGGAACCCCCAAAAAUUAUACAGCAGCCG ......................(((.(((((((((((((.....))))))..))))).)).))). ( -16.30, z-score = -3.81, R) >droSim1.chrX 3573966 65 - 17042790 CACAAAAUCAGAACAUAGCAAAGGCCGCGUGUAUUUUGGAACCACCAAAAAUUAUCCAGCAGCCG ......................(((.((..(((((((((.....))))))..)))...)).))). ( -12.20, z-score = -1.89, R) >droSec1.super_17 1327831 65 + 1527944 CACAAAAUCAGAACAUAGCAACGGCCGCGUGUAUUUUGGAACCACCAAAAAUUAUCCAGCAGCCG .....................((((.((..(((((((((.....))))))..)))...)).)))) ( -13.20, z-score = -2.10, R) >droYak2.chrX 16112499 65 + 21770863 CACAAAAUCAGAAUUUAGAAAAGGCCGCGUGUAUUUUAGAGCCACCGAAGAUAAUACAGCAGCCU .....................((((.((.((((((.................)))))))).)))) ( -11.23, z-score = -1.99, R) >droEre2.scaffold_4690 7822305 65 + 18748788 CACAAAAUCAGAACUUAGAGAAGGCCGCGUGUAUUUUAGAGCCACCGAAGAUUAUACAGCAGCCG ......................(((.(((((((((((.(......).)))).))))).)).))). ( -11.20, z-score = -1.63, R) >droMoj3.scaffold_6473 952479 65 - 16943266 CAGAAAAUCAGUACACUUCAAAGACUAACUGUCGUUAGCAACCGUUAACAGUAAUAAUCCAGACG ...........(((........(((.....)))((((((....)))))).)))............ ( -9.60, z-score = -1.61, R) >consensus CACAAAAUCAGAACAUAGCAAAGGCCGCGUGUAUUUUGGAACCACCAAAAAUUAUACAGCAGCCG ......................(((.(((((((((((((.....))))))..))))).)).))). ( -6.62 = -8.53 + 1.92)
| Location | 17,484,357 – 17,484,466 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 68.72 |
| Shannon entropy | 0.51204 |
| G+C content | 0.56293 |
| Mean single sequence MFE | -37.80 |
| Consensus MFE | -16.30 |
| Energy contribution | -18.18 |
| Covariance contribution | 1.88 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.728328 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
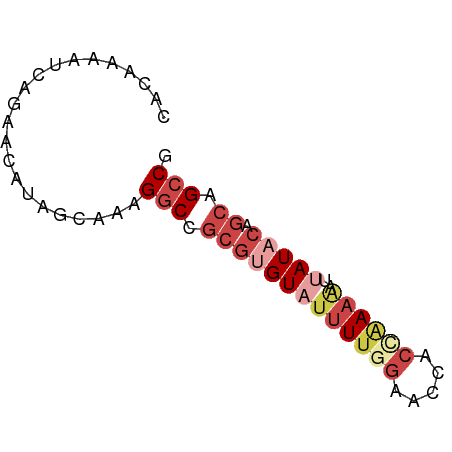
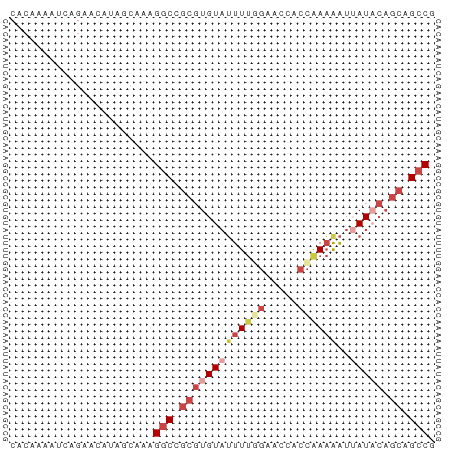
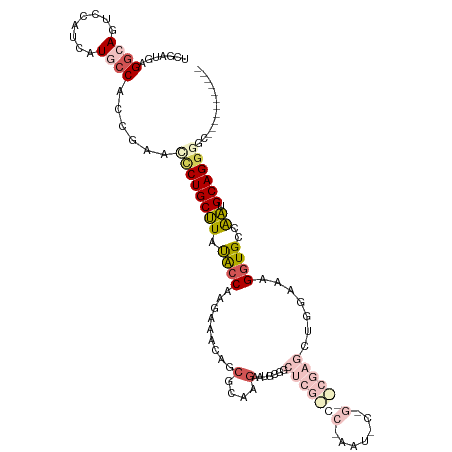
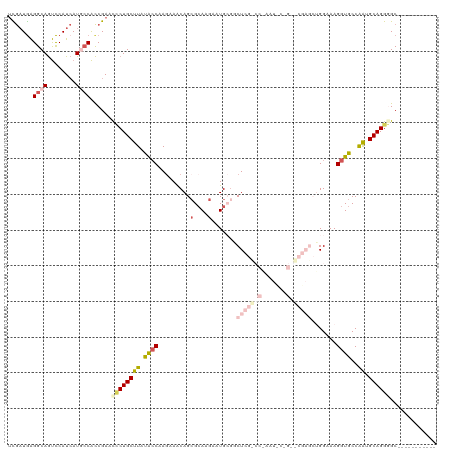
>dm3.chrX 17484357 109 + 22422827 UCGAUGAGGCAGUCCAUCAUUCCACCGAAUCCUGCUUAUAGCAAGAAACAGGGCAAAAAUGCGGCUCGCCCAAAUUCCGAGCGAGCUGGAAAGGUGCCAAUGCAGGGGC----------- ..((((........))))...((.((...((.(((.....))).)).....((((....(.((((((((.(.......).)))))))).)....))))......)))).----------- ( -32.30, z-score = 0.07, R) >droEre2.scaffold_4690 7822370 112 + 18748788 UUGAUAGGGCAGCUCAUCAUAACGCCGAACGCUGCUUACGCCAAGAAGCAGCGGAAGACUGGGGCUCGUCCAAAUCCUGGACGAGCUGGCAAGGUGCCGAUGCAGGUGCUGG-------- .......((((.((((((.....(((...((((((((.(.....))))))))).........((((((((((.....)))))))))))))..(....))))).)).))))..-------- ( -49.20, z-score = -2.93, R) >droYak2.chrX 16112564 120 + 21770863 AUAUUGAGGCAAUCCAUAAUGCCAUCGAACUCUGCUUAUGCCAAAAAGCAGCGCAAGACUGGCACUCGUCCGAAUCCUGGACGAGCUGGUAAGGUGCCGAUGCAGGUGCUCGUUGAGCAC ...((((((((........)))).))))...((((...(((......)))((((...((..((..(((((((.....)))))))))..))...))))....))))((((((...)))))) ( -44.60, z-score = -2.56, R) >droSec1.super_17 1327896 88 + 1527944 UCGAUGAGGCAGUCCAUCAUGCCACCGAACCCUGCCUAUACCAAGAAACAGCGCAAGAAUGCGCCU---------------------GGAAAGGUGCCAGUGCAGGGGC----------- (((....((((........))))..))).((((((((..............(....)...((((((---------------------....)))))).)).))))))..----------- ( -31.60, z-score = -1.95, R) >droSim1.chrX 3574031 88 - 17042790 UCGAUGAGGCAGUCCAUCAUGCCACCGAACCCUGCCUAUACCAAGAAACAGCGCAAGAAUGCGGCU---------------------GGAAAGGUGCCAGUGCAGGGGC----------- (((....((((........))))..))).((((((((.((((......(((((((....))).)))---------------------)....))))..)).))))))..----------- ( -31.30, z-score = -1.55, R) >consensus UCGAUGAGGCAGUCCAUCAUGCCACCGAACCCUGCUUAUACCAAGAAACAGCGCAAGAAUGCGGCUCG_CC_AAU_C_G__CGAGCUGGAAAGGUGCCAAUGCAGGGGC___________ .......((((........))))......((((((((.((((.........(....).......((((((((.....)))))))).......))))..)).))))))............. (-16.30 = -18.18 + 1.88)

| Location | 17,484,357 – 17,484,466 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 68.72 |
| Shannon entropy | 0.51204 |
| G+C content | 0.56293 |
| Mean single sequence MFE | -39.44 |
| Consensus MFE | -19.14 |
| Energy contribution | -21.20 |
| Covariance contribution | 2.06 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.34 |
| SVM RNA-class probability | 0.928684 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
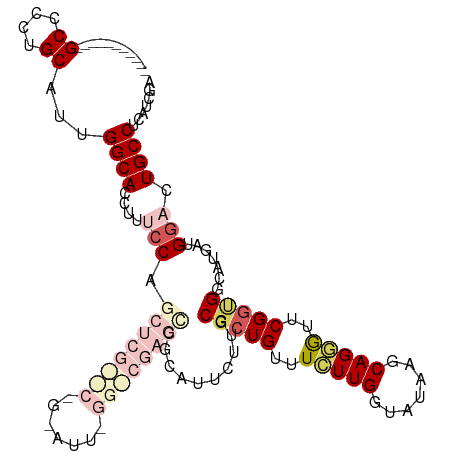
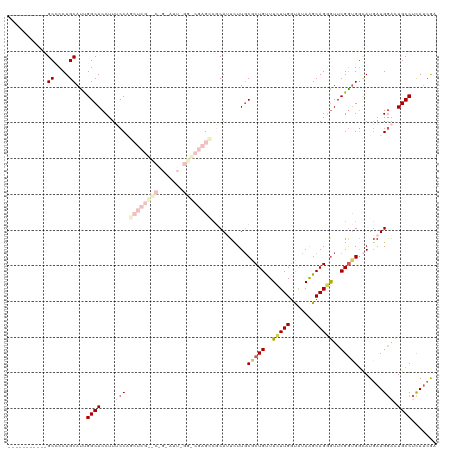
>dm3.chrX 17484357 109 - 22422827 -----------GCCCCUGCAUUGGCACCUUUCCAGCUCGCUCGGAAUUUGGGCGAGCCGCAUUUUUGCCCUGUUUCUUGCUAUAAGCAGGAUUCGGUGGAAUGAUGGACUGCCUCAUCGA -----------(((........)))....((((((((((((((.....))))))))).(((....))).(((..((((((.....))))))..))))))))((((((......)))))). ( -39.60, z-score = -1.64, R) >droEre2.scaffold_4690 7822370 112 - 18748788 --------CCAGCACCUGCAUCGGCACCUUGCCAGCUCGUCCAGGAUUUGGACGAGCCCCAGUCUUCCGCUGCUUCUUGGCGUAAGCAGCGUUCGGCGUUAUGAUGAGCUGCCCUAUCAA --------...(((.((.(((((((.....))).((((((((((...))))))))))....(((...((((((((........))))))))...))).....)))))).)))........ ( -43.70, z-score = -2.38, R) >droYak2.chrX 16112564 120 - 21770863 GUGCUCAACGAGCACCUGCAUCGGCACCUUACCAGCUCGUCCAGGAUUCGGACGAGUGCCAGUCUUGCGCUGCUUUUUGGCAUAAGCAGAGUUCGAUGGCAUUAUGGAUUGCCUCAAUAU ((((((...)))))).......((((.....(((((((((((.......))))))))((((.((..((.((((((........)))))).))..))))))....)))..))))....... ( -45.40, z-score = -2.67, R) >droSec1.super_17 1327896 88 - 1527944 -----------GCCCCUGCACUGGCACCUUUCC---------------------AGGCGCAUUCUUGCGCUGUUUCUUGGUAUAGGCAGGGUUCGGUGGCAUGAUGGACUGCCUCAUCGA -----------..((((((.(((..(((.....---------------------.((((((....)))))).......))).))))))))).(((((((((........))))..))))) ( -35.42, z-score = -1.68, R) >droSim1.chrX 3574031 88 + 17042790 -----------GCCCCUGCACUGGCACCUUUCC---------------------AGCCGCAUUCUUGCGCUGUUUCUUGGUAUAGGCAGGGUUCGGUGGCAUGAUGGACUGCCUCAUCGA -----------..((((((.(((..(((....(---------------------(((.(((....)))))))......))).))))))))).(((((((((........))))..))))) ( -33.10, z-score = -1.17, R) >consensus ___________GCCCCUGCAUUGGCACCUUUCCAGCUCG__C_G_AUU_GG_CGAGCCGCAUUCUUGCGCUGUUUCUUGGUAUAAGCAGGGUUCGGUGGCAUGAUGGACUGCCUCAUCGA ...........((....))...((((........((((((((.......))))))))..((((....(((((..(((((.......)))))..)))))....))))...))))....... (-19.14 = -21.20 + 2.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:53:22 2011