| Sequence ID | dm3.chrX |
|---|---|
| Location | 17,461,429 – 17,461,557 |
| Length | 128 |
| Max. P | 0.803573 |

| Location | 17,461,429 – 17,461,557 |
|---|---|
| Length | 128 |
| Sequences | 12 |
| Columns | 144 |
| Reading direction | reverse |
| Mean pairwise identity | 64.29 |
| Shannon entropy | 0.75928 |
| G+C content | 0.51844 |
| Mean single sequence MFE | -41.25 |
| Consensus MFE | -15.21 |
| Energy contribution | -15.03 |
| Covariance contribution | -0.18 |
| Combinations/Pair | 1.58 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.803573 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
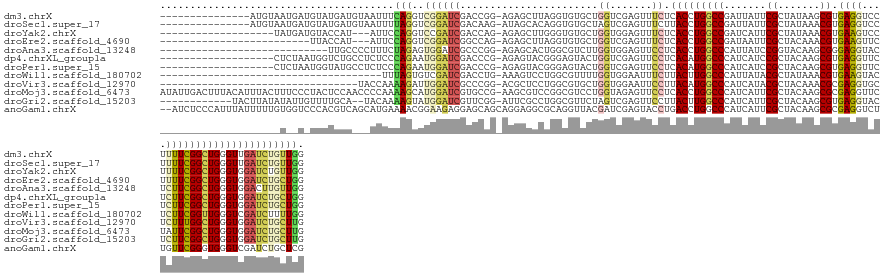
>dm3.chrX 17461429 128 - 22422827 ---------------AUGUAAUGAUGUAUGAUGUAAUUUCAGGUCGGAUCGACCGG-AGAGCUUAGGUGUGCUGGUCGAGUUUCUCACCUGGCCGAUUAUUCGCUAUAAGCGUGAGGUCCUUUUCGGCUGGGUUGAUCUGUUGG ---------------........................(((((((..((((((((-...((....))...)))))))).......(((..(((((...(((((.......))))).......)))))..)))))))))).... ( -41.90, z-score = -1.57, R) >droSec1.super_17 1305075 128 - 1527944 ---------------AUGUAAUGAUGUAUGAUGUAAUUUUAGGUCGGAUCGACAAG-AUAGCACAGGUGUGCUAGUCGAGUUUCUUACCUGGCCGAUUAUUCGCUAUAAACGUGAGGUCCUUUUCGGCUGGGUUGAUCUGUUGG ---------------........................(((((((..(((((...-.((((((....))))))))))).......(((..(((((...(((((.......))))).......)))))..)))))))))).... ( -38.50, z-score = -1.70, R) >droYak2.chrX 16089421 121 - 21770863 -------------------UAUGAUGUACCAU---AUUCCAGGUCCGAUCGACCAG-AGAGCUUGGGUGUGCUGGUGGAGUUUCUCACCUGGCCGAUCAUUCGCUAUAAACGUGAAGUCCUUUUCGGCUGGGUGGAUCUGUUGG -------------------.........(((.---....(((((((..((.(((((-...((....))...))))).)).......(((..(((((...(((((.......))))).......)))))..)))))))))).))) ( -44.00, z-score = -1.94, R) >droEre2.scaffold_4690 7799489 115 - 18748788 -------------------------UUACCAU---AUUCCAGGUCGGAUCGGCCAG-AGAGCUUAGGUGUGCUGGUCGAGUUUCUCACCUGGCCGAUAAUUCGCUACAAACGUGAAGUUCUUUUCGGCUGGGUGGAUCUGCUGG -------------------------.......---...((((..((((((((((((-...((....))...))))))........((((..(((((...(((((.......))))).......)))))..)))))))))))))) ( -46.30, z-score = -3.29, R) >droAna3.scaffold_13248 1866772 115 + 4840945 ----------------------------UUGCCCCUUUCUAGAGUGGAUCGCCCGG-AGAGCACUGGCGUCUUGGUGGAGUUCCUCACCUGGCCCAUUAUCCGGUACAAGCGGGAGGUACUCUUCGGCUGGGUGGACUUGUUGG ----------------------------(..(((.......((.....))(((.((-((((.(((((.(((..(((((......))))).)))))....(((.((....)).)))))).))))))))).)))..)......... ( -38.70, z-score = 1.23, R) >dp4.chrXL_group1a 8981786 124 - 9151740 -------------------CUCUAAUGGUCUGCCUCUCCCAGAAUGGAUCGACCCG-AGAGUACGGGAGUACUGGUCGAGUUCCUCACAUGGCCCAUCAUCCGCUACAAGCGUGAGGUUCUCUUCGGCUGGGUGGAUCUGCUGG -------------------.......((....))....((((...(((((.((((.-..(((((....)))))(((((((..((((((..((........))((.....))))))))..)))...)))))))).))))).)))) ( -47.20, z-score = -1.12, R) >droPer1.super_15 1375098 124 - 2181545 -------------------CUCUAAUGGUAUGCCUCUCCCAGAAUGGAUCGACCCG-AGAGUACGGGAGUACUGGUCGAGUUCCUCACAUGGCCCAUCAUCCGCUACAAGCGUGAGGUUCUCUUCGGCUGGGUGGAUCUGCUGG -------------------.......((....))....((((...(((((.((((.-..(((((....)))))(((((((..((((((..((........))((.....))))))))..)))...)))))))).))))).)))) ( -47.20, z-score = -1.29, R) >droWil1.scaffold_180702 622339 106 - 4511350 -------------------------------------UUUAGUGUCGAUCGACCUG-AAAGUCCUGGCGUUUUGGUGGAAUUUCUUACUUGGCCCAUUAUACGCUAUAAACGUGAAGUACUCUUCGGUUGGGUCGAUCUUUUGG -------------------------------------.........((((((((..-(..((..((((((..((((((...............)))))).))))))...)).(((((....))))).)..))))))))...... ( -31.96, z-score = -2.28, R) >droVir3.scaffold_12970 6214166 110 + 11907090 ---------------------------------UACCAAAAGAUUGGAUCGCCCGG-ACGCUCCUGGCGUGCUGGUGGAAUUCCUUACAUGGCCCAUCAUACGCUACAAACGCGAGGUGCUCUUUGGCUGGGUGGAUCUGCUUG ---------------------------------......(((...(((((((((((-..((.(((.((((((((((((....((......)).))))))...)).....)))).))).)).......)))))).))))).))). ( -35.90, z-score = -0.62, R) >droMoj3.scaffold_6473 15966758 143 - 16943266 AUAUUGACUUUACAUUUACUUUCCCUACUCCAACCCCAAAAGCAUGGAUCGUGCCG-AAGCGUCCGGCGUCCUGGUAGAGUUCCUCACCUGGCCCAUCAUUCGCUACAAGCGCGAGGUUCUAUUCGGCUGGGUGGAUCUGCUUG .......................................(((((.(((.(.(((((-..(((.....)))..)))))..).)))(((((..(((.....(((((.......))))).........)))..)))))...))))). ( -36.64, z-score = 0.52, R) >droGri2.scaffold_15203 7650144 129 - 11997470 ------------UACUUAUAUAUUGUUUUGCA--UACAAAAGUAUGGAUCGUUCGG-AUUCGCCUGGCGUUCUAGUCGAGUUCCUUACUUGGCCCAUCAUUCGCUACAAGCGUGAGGUACUCUUCGGCUGGGUGGAUCUGCUUG ------------..................((--(((....)))))...((..(((-(((((((..((......(((((((.....)))))))((.(((..(((.....)))))))).........))..))))))))))..)) ( -39.30, z-score = -1.94, R) >anoGam1.chrX 6606566 142 + 22145176 --AUCUCCCAUUUAUUUUUGUGGUCCCACGUCAGCAUGAAAACGGAAGAGGAGCAGCAGGAGGCGCAGGUUACGAUCGAGUACCUGACCUGGCCCAUCAUUCGCUACAAGCGCGAGGUCUUGUUCGGGUGGGUCGAUCUGCUCG --..(((((.((..((((..((.(........).))..))))..)).).)))).(((((((((..(((((.((......))))))).)))((((((((...(((.....)))((((......))))))))))))..)))))).. ( -47.40, z-score = -0.25, R) >consensus ____________________U__A_UUAUCAU___AUUCCAGAUUGGAUCGACCAG_AGAGCACGGGCGUGCUGGUCGAGUUCCUCACCUGGCCCAUCAUUCGCUACAAGCGUGAGGUCCUCUUCGGCUGGGUGGAUCUGCUGG .......................................(((..((((((.......................((.......)).(((((((((.......((.......)).((((....)))))))))))))))))))))). (-15.21 = -15.03 + -0.18)
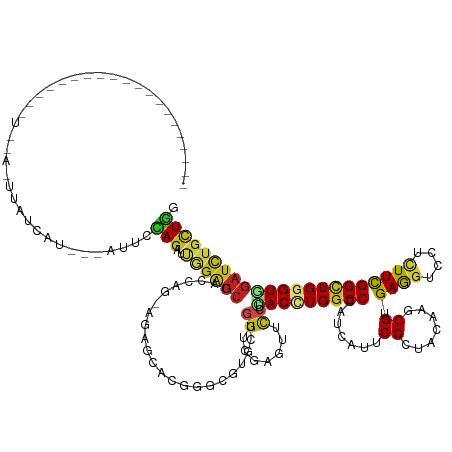

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:53:16 2011