
| Sequence ID | dm3.chrX |
|---|---|
| Location | 17,456,351 – 17,456,441 |
| Length | 90 |
| Max. P | 0.802338 |

| Location | 17,456,351 – 17,456,441 |
|---|---|
| Length | 90 |
| Sequences | 7 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 78.65 |
| Shannon entropy | 0.41539 |
| G+C content | 0.46484 |
| Mean single sequence MFE | -21.06 |
| Consensus MFE | -13.58 |
| Energy contribution | -13.46 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.802338 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 17456351 90 - 22422827 -----CUCCACAUAUUUUAUUAAGUUGGAAACCAGUAAUAAAGUAGGAAGCUCGCUGCCAGCUG-CAGCUCA------CACACACACCUACACGCACACACU-- -----.(((...((((((((((....(....)...))))))))))))).....(((((.....)-))))...------........................-- ( -17.30, z-score = -1.20, R) >droSec1.super_17 1300001 78 - 1527944 -----CUCCACAUAUUUUAUUAAGUUGGAAACCAGUAAUAAAGUAGGAAGCUCGCUGCCAGCUG-CAGCUCA----CACACACACACU---------------- -----.(((...((((((((((....(....)...))))))))))))).....(((((.....)-))))...----............---------------- ( -17.30, z-score = -1.73, R) >droEre2.scaffold_4690 7794348 84 - 18748788 -----CUCCACAUAUUUUAUUAAGUUGGAAACCAGUAAUAAAGUAGGAAGCUCGCUGCCAGCUG-CAGCUCU------CUCACACACACACACUCU-------- -----.(((...((((((((((....(....)...))))))))))))).....(((((.....)-))))...------..................-------- ( -17.30, z-score = -1.44, R) >droYak2.chrX 16084577 98 - 21770863 -----CUCCACAUAUUUUAUUAAGUUGGAAACCAGUAAUAAAGUAGGAAGCUCGUUGCCAGCUG-CAGCUCACAGACGCACACACACCUACACUCACACACACU -----..........(((((((....(....)...)))))))(((((.((((.......)))).-..((........)).......)))))............. ( -15.60, z-score = -0.24, R) >droAna3.scaffold_13248 1859485 100 + 4840945 CGACGCUCCACAUAUUUUAUUAAGUUGGAAACCAGUAAUAAAGUAGGAAGCCCGCUGCCCGCUGCCAGCUGCAGCUCACCAACACACUGCCACGCACACA---- ....(((((...((((((((((....(....)...)))))))))))).)))..(((((..((.....)).))))).........................---- ( -21.80, z-score = -0.26, R) >dp4.chrXL_group1a 8976373 96 - 9151740 CGACGCUCCACAUAUUUUAUUAAGCUGGAAACCAGUAAUAAAGUAGGAAGCUCGCUGCCAGCUG-CAGCUCG----CUCUCACAAACCGUCGAAAGAGAAA--- (((((.......(((((((((..(((((...))))))))))))))(..(((..(((((.....)-))))..)----))..)......))))).........--- ( -29.40, z-score = -2.90, R) >droPer1.super_15 1369562 95 - 2181545 CGACGCUCCACAUAUUUUAUUAAGCUGGAAACCAGUAAUAAAGUAGGAAGCUCGCUGCCAGCUG-CAGCUCG----CUCUCACAAACCGUCGCACAGAAA---- (((((.......(((((((((..(((((...))))))))))))))(..(((..(((((.....)-))))..)----))..)......)))))........---- ( -28.70, z-score = -2.84, R) >consensus _____CUCCACAUAUUUUAUUAAGUUGGAAACCAGUAAUAAAGUAGGAAGCUCGCUGCCAGCUG_CAGCUCA____C_CACACACACCGACACACACACA____ ......(((...((((((((((....(....)...))))))))))))).....((((........))))................................... (-13.58 = -13.46 + -0.12)

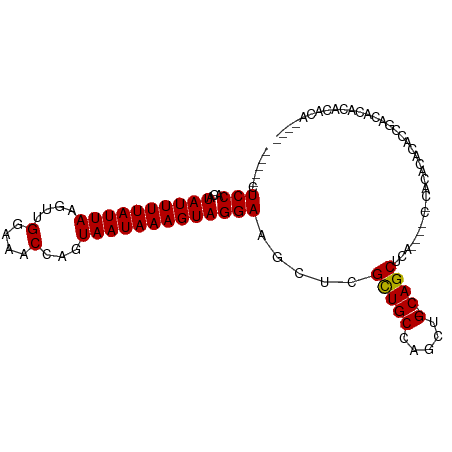
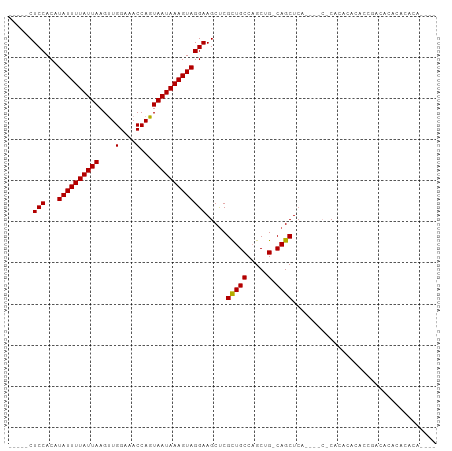
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:53:15 2011