
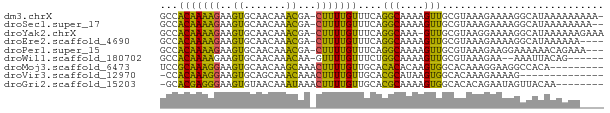
| Sequence ID | dm3.chrX |
|---|---|
| Location | 17,446,520 – 17,446,592 |
| Length | 72 |
| Max. P | 0.520573 |

| Location | 17,446,520 – 17,446,592 |
|---|---|
| Length | 72 |
| Sequences | 9 |
| Columns | 74 |
| Reading direction | forward |
| Mean pairwise identity | 75.38 |
| Shannon entropy | 0.49840 |
| G+C content | 0.38473 |
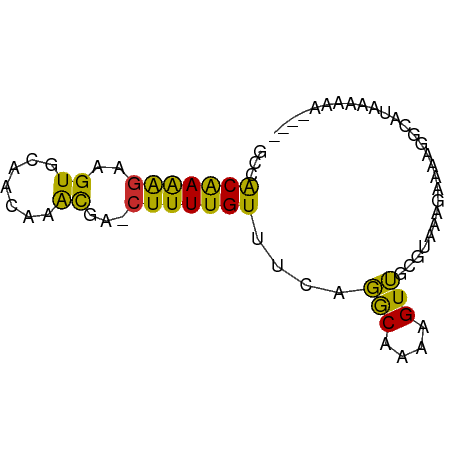
| Mean single sequence MFE | -14.98 |
| Consensus MFE | -6.08 |
| Energy contribution | -4.80 |
| Covariance contribution | -1.28 |
| Combinations/Pair | 1.54 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.520573 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chrX 17446520 72 + 22422827 GCCACAAAAGAAGUGCAACAAACGA-CUUUUGUUUCAGGCAAAAGUUGCGUAAAGAAAAGGCAUAAAAAAAAA- ((((((((((..((.......))..-))))))).....(((.....)))..........)))...........- ( -14.50, z-score = -1.83, R) >droSec1.super_17 1292092 71 + 1527944 GCCACAAAAGAAGUGCAACAAACGA-CUUUUGUUUCAGGCAAAAGUUGCGUAAAGAAAAGGCAUAAAAAAAA-- ((((((((((..((.......))..-))))))).....(((.....)))..........)))..........-- ( -14.50, z-score = -1.78, R) >droYak2.chrX 16075464 72 + 21770863 GCCACAAAAGAAGUGCAACAAACGA-CUUUUGUUUCAGGCAAA-GUUGCGUAAGGAAAAGGCAUAAAAAAGAAA ((((((((((..((.......))..-))))))).....(((..-..)))..........)))............ ( -14.50, z-score = -1.42, R) >droEre2.scaffold_4690 7785372 69 + 18748788 GCCACAAAAGAAGUGCAACAAACGA-CUUUUGUUUCAGGCAAAAGUUGCGUAAAGAAAAGGCAUAAAAAA---- ((((((((((..((.......))..-))))))).....(((.....)))..........)))........---- ( -14.50, z-score = -1.63, R) >droPer1.super_15 1358432 70 + 2181545 GCCACAAAAGAAGUGCAACAAACGA-CUUUUGUUUCAGGCAAAAGUUGCGUAAAGAAGGAAAAAACAGAAA--- ((.((.......))))......(((-(((((((.....)))))))))).......................--- ( -11.40, z-score = -0.34, R) >droWil1.scaffold_180702 609372 65 + 4511350 GCCACAAAAGAAGUGCAACAAACAA-GUUUUGUUUCUGGCAAAAGUUGCGUAAAGAA--AAAUUACAG------ ((.((.......))))......(((-.((((((.....)))))).))).((((....--...))))..------ ( -8.60, z-score = 0.52, R) >droMoj3.scaffold_6473 15947902 65 + 16943266 UCCGCAAAGGAAGUGCAACAAGCAAACUUUUGUUGCACACACAAGUGGCACAAAGGAAGGCCACA--------- (((.....))).((((((((((......))))))))))......(((((.(.......)))))).--------- ( -23.80, z-score = -3.77, R) >droVir3.scaffold_12970 6196424 59 - 11907090 -CCACAAAGGAAGUGCAGCAAACAAACUUUUGUUGCACGCAUAAGUGGCACAAAGAAAAG-------------- -((((.......((((((((((......))))))))))......))))............-------------- ( -17.42, z-score = -3.20, R) >droGri2.scaffold_15203 7631970 65 + 11997470 -GCACGAGGGAAGUGUAACAAAUAAACUUUUGUUGCACGCAAAAGUGGCACACAGAAUAGUUACAA-------- -((.(....)..((((((((((......))))))))))))....(((((..........)))))..-------- ( -15.60, z-score = -2.01, R) >consensus GCCACAAAAGAAGUGCAACAAACGA_CUUUUGUUUCAGGCAAAAGUUGCGUAAAGAAAAGGCAUAAAAAA____ ............((((((((((......))))))...(((....)))))))....................... ( -6.08 = -4.80 + -1.28)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:53:13 2011