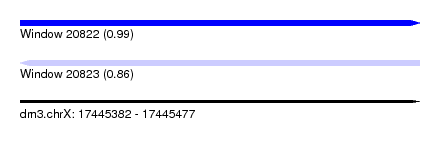
| Sequence ID | dm3.chrX |
|---|---|
| Location | 17,445,382 – 17,445,477 |
| Length | 95 |
| Max. P | 0.994991 |

| Location | 17,445,382 – 17,445,477 |
|---|---|
| Length | 95 |
| Sequences | 7 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 68.79 |
| Shannon entropy | 0.59455 |
| G+C content | 0.50946 |
| Mean single sequence MFE | -33.31 |
| Consensus MFE | -21.13 |
| Energy contribution | -22.77 |
| Covariance contribution | 1.64 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.75 |
| SVM RNA-class probability | 0.994991 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
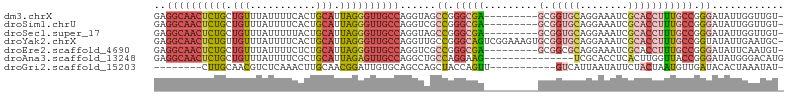
>dm3.chrX 17445382 95 + 22422827 GAGGCAACUCUGCUGUUUAUUUUCACUGCAUUAGGGUUGCCAGGUAGCCGGGCGA---------GCGGUGCAGGAAAUCGCACCUUUGCCGGGAUAUUGGUUGU- ..((((((((((.(((...........))).)))))))))).((((.((.(((((---------(.(((((........))))))))))).)).))))......- ( -40.10, z-score = -3.03, R) >droSim1.chrU 5270402 95 - 15797150 GAGGCAACUCUGCUGUUUAUUUUCACUGCAUUAGGGUUGCCAGGUCGCCGGGCGA---------GCGGUGCAGGAAAUCGCACCUUUGCCGGGAUAUUGGUUGU- ..((((((((((.(((...........))).))))))))))......((.(((((---------(.(((((........))))))))))).))...........- ( -38.80, z-score = -2.44, R) >droSec1.super_17 1290934 95 + 1527944 GAGGCAACUCUGCUGUUUAUUUUUACUGCAUUAGGGUUGCCAGGUAGCCGGGCGA---------GCGGUGCAGGAAAUCGCACCUUUGCCGGGAUAUUGGUUGU- ..((((((((((.(((...........))).)))))))))).((((.((.(((((---------(.(((((........))))))))))).)).))))......- ( -40.10, z-score = -3.22, R) >droYak2.chrX 16074218 104 + 21770863 GAGGCAACUCUGUUGUUUAUUUUCACUGCAUUAGGGUUGCCAGGUUGCCGGGCAGUCGGAAAGUGCGGUGCAGGAAAUCGCACCUUUGCCGGUAUAUUGAAUGC- ..((((((((((.(((...........))).))))))))))....(((((((((.(.....).)))(((((........)))))....))))))..........- ( -37.90, z-score = -2.06, R) >droEre2.scaffold_4690 7784306 95 + 18748788 GAGGCAACUCUGCUGUUUAUUUUCUCUGCAUUAGGGUUGCCAGGUCGCCGGGCGA---------GCGGCGCAGGAAAUCGCACCUUUGCCGGGAUAUUCAAUGU- ..((((((((((.(((...........))).)))))))))).((...((.(((((---------(.((.((........)).)))))))).))....)).....- ( -35.40, z-score = -1.44, R) >droAna3.scaffold_13248 1852017 90 - 4840945 GAGGCAACUCUGCUGUUUAUUUUCGCUGCAUUAGAGUUGCCAGGCUGCCAGGAAG---------------UCGCACCUCACUUGGUUACCGGGAUAUGGGACAUG ..((((((((((.(((...........))).)))))))))).((..((((((.((---------------......))..))))))..))............... ( -28.10, z-score = -0.68, R) >droGri2.scaffold_15203 7630353 85 + 11997470 --------CUUGCAACGUCUCAAACUUGCAACGGAUUGUGCAGCCAGCUACCAGUU-----------GUCAUUAAUAUUCUACUAAUGUUGAUACACUAAAUAU- --------.((((((.((.....))))))))......(((..(.((((.....)))-----------).)(((((((((.....))))))))).))).......- ( -12.80, z-score = 0.35, R) >consensus GAGGCAACUCUGCUGUUUAUUUUCACUGCAUUAGGGUUGCCAGGUAGCCGGGCGA_________GCGGUGCAGGAAAUCGCACCUUUGCCGGGAUAUUGGAUGU_ ..((((((((((.(((...........))).))))))))))......((.(((((...........(((((........))))).))))).))............ (-21.13 = -22.77 + 1.64)
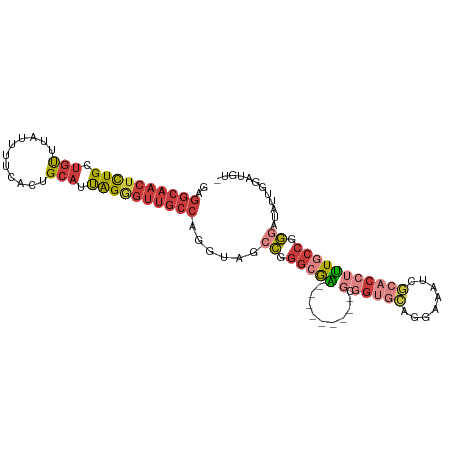
| Location | 17,445,382 – 17,445,477 |
|---|---|
| Length | 95 |
| Sequences | 7 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 68.79 |
| Shannon entropy | 0.59455 |
| G+C content | 0.50946 |
| Mean single sequence MFE | -25.78 |
| Consensus MFE | -12.27 |
| Energy contribution | -13.60 |
| Covariance contribution | 1.33 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.862355 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 17445382 95 - 22422827 -ACAACCAAUAUCCCGGCAAAGGUGCGAUUUCCUGCACCGC---------UCGCCCGGCUACCUGGCAACCCUAAUGCAGUGAAAAUAAACAGCAGAGUUGCCUC -..............((((..((((((......))))))((---------(((((.((...)).)))........(((.((........)).))))))))))).. ( -28.70, z-score = -2.29, R) >droSim1.chrU 5270402 95 + 15797150 -ACAACCAAUAUCCCGGCAAAGGUGCGAUUUCCUGCACCGC---------UCGCCCGGCGACCUGGCAACCCUAAUGCAGUGAAAAUAAACAGCAGAGUUGCCUC -..........(((((((...((((((......))))))..---------..).)))).))...((((((.....(((.((........)).)))..)))))).. ( -28.90, z-score = -2.08, R) >droSec1.super_17 1290934 95 - 1527944 -ACAACCAAUAUCCCGGCAAAGGUGCGAUUUCCUGCACCGC---------UCGCCCGGCUACCUGGCAACCCUAAUGCAGUAAAAAUAAACAGCAGAGUUGCCUC -..............((((..((((((......))))))((---------(((((.((...)).)))........(((.((........)).))))))))))).. ( -27.80, z-score = -2.34, R) >droYak2.chrX 16074218 104 - 21770863 -GCAUUCAAUAUACCGGCAAAGGUGCGAUUUCCUGCACCGCACUUUCCGACUGCCCGGCAACCUGGCAACCCUAAUGCAGUGAAAAUAAACAACAGAGUUGCCUC -..............((((((((((((...........)))))))).....)))).((((((((((((.......))).((........))..))).)))))).. ( -29.30, z-score = -2.22, R) >droEre2.scaffold_4690 7784306 95 - 18748788 -ACAUUGAAUAUCCCGGCAAAGGUGCGAUUUCCUGCGCCGC---------UCGCCCGGCGACCUGGCAACCCUAAUGCAGAGAAAAUAAACAGCAGAGUUGCCUC -..........(((((((...((((((......))))))..---------..).)))).))...((((((.....(((..............)))..)))))).. ( -26.24, z-score = -0.63, R) >droAna3.scaffold_13248 1852017 90 + 4840945 CAUGUCCCAUAUCCCGGUAACCAAGUGAGGUGCGA---------------CUUCCUGGCAGCCUGGCAACUCUAAUGCAGCGAAAAUAAACAGCAGAGUUGCCUC ...............(((..(((.(.((((.....---------------))))))))..))).(((((((((..((.............))..))))))))).. ( -24.62, z-score = -0.87, R) >droGri2.scaffold_15203 7630353 85 - 11997470 -AUAUUUAGUGUAUCAACAUUAGUAGAAUAUUAAUGAC-----------AACUGGUAGCUGGCUGCACAAUCCGUUGCAAGUUUGAGACGUUGCAAG-------- -.......((((((((.((((((((...))))))))((-----------.....))...))).)))))......(((((((((...))).)))))).-------- ( -14.90, z-score = 0.94, R) >consensus _ACAUCCAAUAUCCCGGCAAAGGUGCGAUUUCCUGCACCGC_________UCGCCCGGCUACCUGGCAACCCUAAUGCAGUGAAAAUAAACAGCAGAGUUGCCUC ...............(((((.((((((......)))))).............(((.((...)).)))...............................))))).. (-12.27 = -13.60 + 1.33)

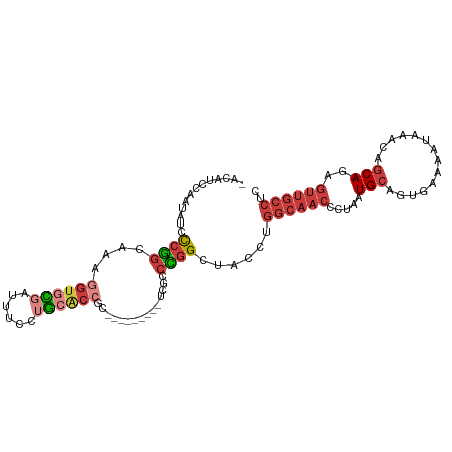
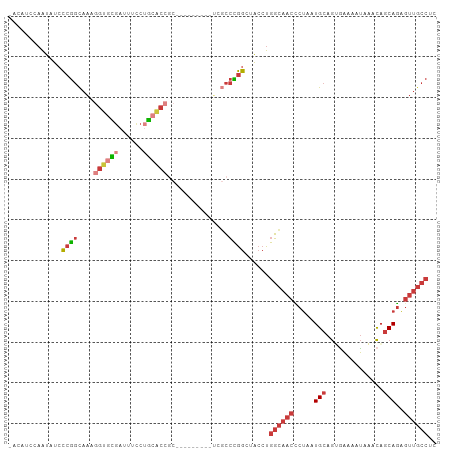
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:53:12 2011