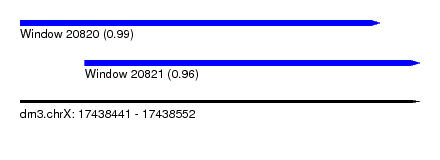
| Sequence ID | dm3.chrX |
|---|---|
| Location | 17,438,441 – 17,438,552 |
| Length | 111 |
| Max. P | 0.991449 |

| Location | 17,438,441 – 17,438,541 |
|---|---|
| Length | 100 |
| Sequences | 7 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 73.80 |
| Shannon entropy | 0.50451 |
| G+C content | 0.49272 |
| Mean single sequence MFE | -31.10 |
| Consensus MFE | -20.87 |
| Energy contribution | -21.80 |
| Covariance contribution | 0.93 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.48 |
| SVM RNA-class probability | 0.991449 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
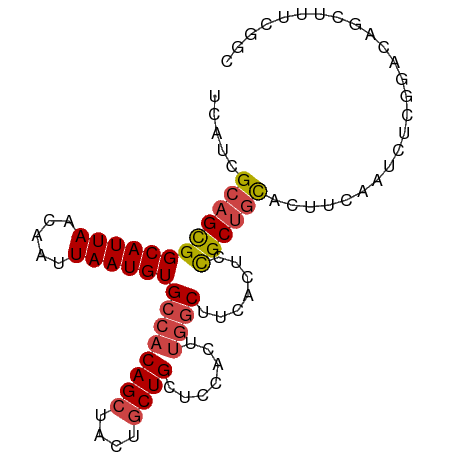
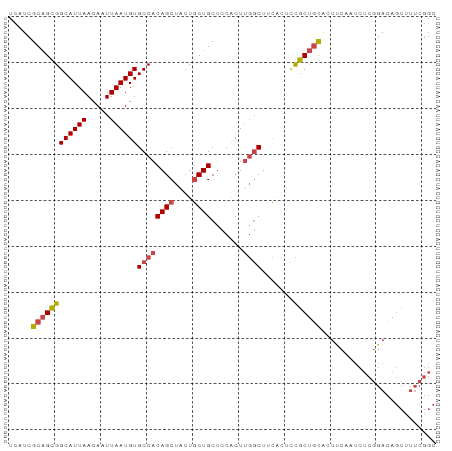
>dm3.chrX 17438441 100 + 22422827 UUGCAAUUGCAGCUGAAGUCA-----UCGCAGCGGCAUUAA--CAAUUAAUGUGCCACAGCU-ACUGCUGCUCCACUUGGCUUCACUUCGCUGCACUUCAAUCUCGGA-- (((.((.((((((((((((((-----..(.((((((((...--........)))))..(((.-...)))))).)...))))))))....)))))).))))).......-- ( -31.50, z-score = -2.20, R) >droEre2.scaffold_4690 7777422 100 + 18748788 UUGCAAUUGCAGCUGAAGUCA-----UCGCAGCGGCAUUAA--CAAUUAAUGUGCCACAGCU-ACUGCUGCUCCACUUGGCUUCACUUCGCUGCAAUCCAAUCUCGGA-- ......(((((((((((((((-----..(.((((((((...--........)))))..(((.-...)))))).)...))))))))....)))))))(((......)))-- ( -33.50, z-score = -2.83, R) >droYak2.chrX 16067204 100 + 21770863 UUGCAGUUGCAGCUGAAGUCA-----UCGCAGCGGCAUUAA--CAAUUAAUGUGCCACAGCU-ACUGCUGCUCCACUUGGCUUCACUCCGCUGCACUUCAAUCUCGGA-- (((.(((.(((((((((((((-----..(.((((((((...--........)))))..(((.-...)))))).)...))))))))....)))))))).))).......-- ( -33.10, z-score = -2.11, R) >droSec1.super_17 1284013 100 + 1527944 UUGCAAUUGCAGCUGAAGUCA-----UCGCAGCGGCAUUAA--CAAUUAAUGUGCCACAGCU-ACUGCUGCUCCACUUGGCUUCACUCCGCUGCACUUCAAUCUCGGA-- (((.((.((((((((((((((-----..(.((((((((...--........)))))..(((.-...)))))).)...))))))))....)))))).))))).......-- ( -31.50, z-score = -2.18, R) >droSim1.chrX_random 4461056 100 + 5698898 UUGCAAUUGCAGCUGAAGUCA-----UCGCAGCGGCAUUAA--CAAUUAAUGUGCCACAGCU-ACUGCUGCUCCACUUGGCUUCACUCCGCUGCACUUCAAUCUCGGA-- (((.((.((((((((((((((-----..(.((((((((...--........)))))..(((.-...)))))).)...))))))))....)))))).))))).......-- ( -31.50, z-score = -2.18, R) >droPer1.super_15 1348325 95 + 2181545 ------CCGAAGUUGAAGUCA-----UUGCUGUGGCAUUAA--CAAUUAAUGUGCCACAGC--GCUACUGCGCUGCUCCACUUUGGCAUGCCAUGCCACGCCACCCAAUC ------..(((((.(((((..-----..((((((((((...--........))))))))))--((....)))))..)).)))))(((((...)))))............. ( -29.60, z-score = -1.69, R) >droGri2.scaffold_15110 2573597 104 - 24565398 CUGCAGCUGCAACUGAAAUUCGUGACUUGAAGCGACAUUAAGCUAGGGGAUUUAGUAUGGCCAAAGGCAUUUCCAAAUGCCUUUGCUUUA----AUUGCACCCUCAAA-- .(((((((...((((((.(((.((.(((((.......))))).)).))).))))))..)))((((((((((....)))))))))).....----..))))........-- ( -27.00, z-score = -0.60, R) >consensus UUGCAAUUGCAGCUGAAGUCA_____UCGCAGCGGCAUUAA__CAAUUAAUGUGCCACAGCU_ACUGCUGCUCCACUUGGCUUCACUCCGCUGCACUUCAAUCUCGGA__ .......((((((((((((((.......((((((((((.............)))))..((....)))))))......))))))))....))))))............... (-20.87 = -21.80 + 0.93)
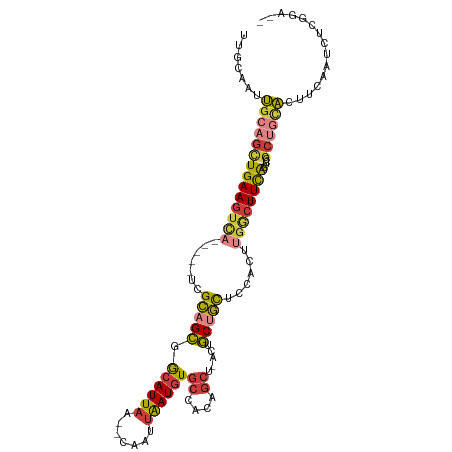
| Location | 17,438,459 – 17,438,552 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 83.58 |
| Shannon entropy | 0.32219 |
| G+C content | 0.52384 |
| Mean single sequence MFE | -26.54 |
| Consensus MFE | -19.19 |
| Energy contribution | -19.78 |
| Covariance contribution | 0.58 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.72 |
| SVM RNA-class probability | 0.963618 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 17438459 93 + 22422827 UCAUCGCAGCGGCAUUAACAAUUAAUGUGCCACAGCUACUGCUGCUCCACUUGGCUUCACUUCGCUGCACUUCAAUCUCGGACAGCUUUCGGC .....((((((((((((.....))))))((((((((....)))).......)))).......)))))).........(((((.....))))). ( -25.71, z-score = -2.08, R) >droEre2.scaffold_4690 7777440 93 + 18748788 UCAUCGCAGCGGCAUUAACAAUUAAUGUGCCACAGCUACUGCUGCUCCACUUGGCUUCACUUCGCUGCAAUCCAAUCUCGGACAGCUUUCGGC .....((((((((((((.....))))))((((((((....)))).......)))).......))))))..(((......)))........... ( -26.41, z-score = -2.16, R) >droYak2.chrX 16067222 93 + 21770863 UCAUCGCAGCGGCAUUAACAAUUAAUGUGCCACAGCUACUGCUGCUCCACUUGGCUUCACUCCGCUGCACUUCAAUCUCGGACAGCUUUCGGC .....((((((((((((.....))))))((((((((....)))).......)))).......)))))).........(((((.....))))). ( -25.91, z-score = -2.02, R) >droSec1.super_17 1284031 93 + 1527944 UCAUCGCAGCGGCAUUAACAAUUAAUGUGCCACAGCUACUGCUGCUCCACUUGGCUUCACUCCGCUGCACUUCAAUCUCGGACAGCAUUCGGC .....((((((((((((.....))))))((((((((....)))).......)))).......)))))).........(((((.....))))). ( -25.81, z-score = -1.93, R) >droSim1.chrX_random 4461074 93 + 5698898 UCAUCGCAGCGGCAUUAACAAUUAAUGUGCCACAGCUACUGCUGCUCCACUUGGCUUCACUCCGCUGCACUUCAAUCUCGGACAGCUUUCGGC .....((((((((((((.....))))))((((((((....)))).......)))).......)))))).........(((((.....))))). ( -25.91, z-score = -2.02, R) >droPer1.super_15 1348337 83 + 2181545 UCAUUGCUGUGGCAUUAACAAUUAAUGUGCCACAGC-GCUACUGCGCUGCUCCACUUUGGCAUGCCAUGCCACGCCACCCAAUC--------- ..((((..(((((.............(((...((((-((....))))))...)))..((((((...)))))).))))).)))).--------- ( -29.50, z-score = -3.32, R) >consensus UCAUCGCAGCGGCAUUAACAAUUAAUGUGCCACAGCUACUGCUGCUCCACUUGGCUUCACUCCGCUGCACUUCAAUCUCGGACAGCUUUCGGC .....((((((((((((.....))))))((((((((....)))).......)))).......))))))......................... (-19.19 = -19.78 + 0.58)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:53:11 2011