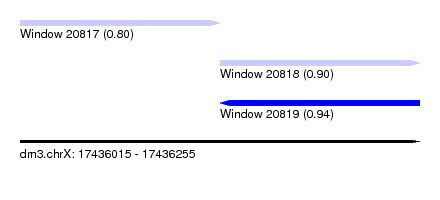
| Sequence ID | dm3.chrX |
|---|---|
| Location | 17,436,015 – 17,436,255 |
| Length | 240 |
| Max. P | 0.937520 |

| Location | 17,436,015 – 17,436,135 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.00 |
| Shannon entropy | 0.32617 |
| G+C content | 0.40833 |
| Mean single sequence MFE | -25.65 |
| Consensus MFE | -14.91 |
| Energy contribution | -16.73 |
| Covariance contribution | 1.81 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.799816 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 17436015 120 + 22422827 UCUUUUACUUAUAAAUUGCUGGAAUUUUCCGCAGAUUUUCUUCGCACUCAGUCAAGUGUGAAGCGAAUGGCACACCAUUUCCUCCCGCUUCUCUUCUCCCAAGAUUACAGAGACUUGUCU ...............((((.(((....))))))).....(((((((((......))))))))).((((((....)))))).........((((((((....)))....)))))....... ( -24.50, z-score = -0.98, R) >droEre2.scaffold_4690 7774961 96 + 18748788 UCUUUC-CUUAUAAAUUGCUCGAACUUUCCGCAGAUUUUUGC-----------------------AAUGCCACAACAUUUUCCCCAUCUUCUCUACUACAAAGAUCACAGAGACUUGUUU ......-.......(((((..(((((......)).)))..))-----------------------))).....((((.((((...(((((..........)))))....))))..)))). ( -7.60, z-score = 0.40, R) >droSec1.super_17 1281446 120 + 1527944 UCUUUCAUUUAUAAAUUGCUGGAAUUUUCCGCAGAUUUUCUUCACACUCAGUCAAGUGUGAAGCGAAUGGCACACCAUUUCCUUCGUCUUCUCUUCUCCGAAGAUCACAGCGACUUGUCU ..........((((.((((((..((((((.(.(((....(((((((((......))))))))).((((((....))))))..............)))).))))))..)))))).)))).. ( -32.20, z-score = -3.38, R) >droSim1.chrX_random 4458620 120 + 5698898 UCUUUCAUUUAUAAAUUGCUGGAAUUUUCCGGAGAUUUUCUUCACACUCAGUCAAGUGUGAAGCGAAUGGCACACCAUUUCCUUCGUCUUCUCUUCUCCGAAGAUCACAGCGACUUGUCU ..........((((.((((((..((((((.(((((....(((((((((......))))))))).((((((....))))))..............)))))))))))..)))))).)))).. ( -38.30, z-score = -4.77, R) >consensus UCUUUCACUUAUAAAUUGCUGGAAUUUUCCGCAGAUUUUCUUCACACUCAGUCAAGUGUGAAGCGAAUGGCACACCAUUUCCUCCGUCUUCUCUUCUCCGAAGAUCACAGAGACUUGUCU ..........((((.((((((((....)))...(((((((((((((((......))))))))..((((((....))))))...................)))))))..))))).)))).. (-14.91 = -16.73 + 1.81)

| Location | 17,436,135 – 17,436,255 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.58 |
| Shannon entropy | 0.16684 |
| G+C content | 0.45985 |
| Mean single sequence MFE | -38.02 |
| Consensus MFE | -31.96 |
| Energy contribution | -32.44 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.84 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.895757 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 17436135 120 + 22422827 GCUAAUUGUAGCCUGCGGAGUUGAAUUGCAGGGUUAGAUGGGUUUAUCUAACACCUGUCACUUGAUUGUCAAUCUGACACGCGAUCGAGGGAAGCGUCAUUAAGCAGACUUCAGGUUUCG .......(.((((((..(((((.....(((((((((((((....)))))))).)))))..(((((((((...........)))))))))....((........)).))))))))))).). ( -35.70, z-score = -1.24, R) >droEre2.scaffold_4690 7775057 120 + 18748788 GCUAAUUGUUGCUUGUGGAGUUGGAUUGCCGGGUUGGAUGGUUUUAUCUAACACCUGUCAUUUGAUUGUCAAUCUGACACGCGAUCAAGGGAAGCGUCAUUAAAACGACUUCAGGCUCCG ((........))....((((((.((((((.((((((((((....)))))))).))(((((.(((.....)))..))))).))))))....(((((((.......))).)))).)))))). ( -37.40, z-score = -2.00, R) >droYak2.chrX 16064648 119 + 21770863 GCUAAUUGUAGCUCGAGGUGUUGGAUUGCAGGGUUAGAUGGGUUUAUCUAACACCUGUCACUUGAUUGUCAAUCUGACACGCGAUCGCGAGAAGCGUCAUUAAAACGACUUUAGGUGCC- (((..(((..(.(((..((((..(((((((((((((((((....)))))))).))))((....))....)))))..)))).))))..)))..)))(((........)))..........- ( -38.00, z-score = -2.17, R) >droSec1.super_17 1281566 120 + 1527944 GCUAAUUGUAGCCUGAGGAGUUGGAUUGCAGGGUUAGAUGGGUUUAUCUAACACCUGUCACUUGAUUGUCACUCUGACACGCGAUCGAGGGUAGCGUCAUUAAGAAGACUUCAGGUUUCG .......(.(((((((((.........(((((((((((((....)))))))).)))))..((((((.(((.....)))((((.((.....)).)))).))))))....))))))))).). ( -38.10, z-score = -1.79, R) >droSim1.chrX_random 4458740 119 + 5698898 GCUAAUUGUAGCCUGAGGAGUUGGAUUGCAGGGUUAGAUGGGUUUAUCUAACACCUGUCACUUGAUUGUCAAUCUGACACGCGAUCGAGGGUAGCGUCAUUAAGAAGACUUCAGGUUAA- ........((((((((((.((..(((((((((((((((((....)))))))).))))((....))....)))))..))((((.((.....)).))))...........)))))))))).- ( -40.90, z-score = -3.13, R) >consensus GCUAAUUGUAGCCUGAGGAGUUGGAUUGCAGGGUUAGAUGGGUUUAUCUAACACCUGUCACUUGAUUGUCAAUCUGACACGCGAUCGAGGGAAGCGUCAUUAAGAAGACUUCAGGUUCCG ..........((((((((.(((((((((((((((((((((....)))))))).))))((....))....)))))))))((((...(....)..))))...........)))))))).... (-31.96 = -32.44 + 0.48)

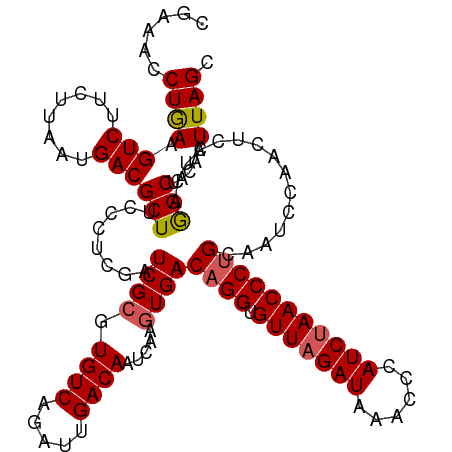
| Location | 17,436,135 – 17,436,255 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.58 |
| Shannon entropy | 0.16684 |
| G+C content | 0.45985 |
| Mean single sequence MFE | -30.71 |
| Consensus MFE | -23.68 |
| Energy contribution | -23.88 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.937520 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 17436135 120 - 22422827 CGAAACCUGAAGUCUGCUUAAUGACGCUUCCCUCGAUCGCGUGUCAGAUUGACAAUCAAGUGACAGGUGUUAGAUAAACCCAUCUAACCCUGCAAUUCAACUCCGCAGGCUACAAUUAGC .......((.(((((((.......(((((...((((((........)))))).....))))).((((.(((((((......)))))))))))............))))))).))...... ( -33.80, z-score = -3.21, R) >droEre2.scaffold_4690 7775057 120 - 18748788 CGGAGCCUGAAGUCGUUUUAAUGACGCUUCCCUUGAUCGCGUGUCAGAUUGACAAUCAAAUGACAGGUGUUAGAUAAAACCAUCCAACCCGGCAAUCCAACUCCACAAGCAACAAUUAGC .((((...((((.((((.....))))))))....(((.((.(((((..(((.....))).)))))((.(((.(((......))).))))).)).)))...))))................ ( -28.10, z-score = -1.55, R) >droYak2.chrX 16064648 119 - 21770863 -GGCACCUAAAGUCGUUUUAAUGACGCUUCUCGCGAUCGCGUGUCAGAUUGACAAUCAAGUGACAGGUGUUAGAUAAACCCAUCUAACCCUGCAAUCCAACACCUCGAGCUACAAUUAGC -.....((((.(((((....)))))((((...((....))((((..(((((((......))..((((.(((((((......))))))))))))))))..))))...)))).....)))). ( -31.40, z-score = -2.57, R) >droSec1.super_17 1281566 120 - 1527944 CGAAACCUGAAGUCUUCUUAAUGACGCUACCCUCGAUCGCGUGUCAGAGUGACAAUCAAGUGACAGGUGUUAGAUAAACCCAUCUAACCCUGCAAUCCAACUCCUCAGGCUACAAUUAGC .....(((((.(((..(((..((((((.............))))))))).))).......((.((((.(((((((......)))))))))))))..........)))))........... ( -30.82, z-score = -2.63, R) >droSim1.chrX_random 4458740 119 - 5698898 -UUAACCUGAAGUCUUCUUAAUGACGCUACCCUCGAUCGCGUGUCAGAUUGACAAUCAAGUGACAGGUGUUAGAUAAACCCAUCUAACCCUGCAAUCCAACUCCUCAGGCUACAAUUAGC -....(((((.(((.......((((((.............))))))....))).......((.((((.(((((((......)))))))))))))..........)))))........... ( -29.42, z-score = -2.84, R) >consensus CGAAACCUGAAGUCUUCUUAAUGACGCUUCCCUCGAUCGCGUGUCAGAUUGACAAUCAAGUGACAGGUGUUAGAUAAACCCAUCUAACCCUGCAAUCCAACUCCUCAGGCUACAAUUAGC ......((((.(((........)))(((........((((.((((.....)))).....))))((((.(((((((......)))))))))))...............))).....)))). (-23.68 = -23.88 + 0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:53:09 2011