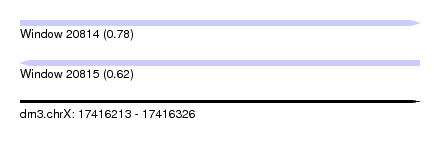
| Sequence ID | dm3.chrX |
|---|---|
| Location | 17,416,213 – 17,416,326 |
| Length | 113 |
| Max. P | 0.783300 |

| Location | 17,416,213 – 17,416,326 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 71.72 |
| Shannon entropy | 0.45825 |
| G+C content | 0.40288 |
| Mean single sequence MFE | -26.89 |
| Consensus MFE | -12.24 |
| Energy contribution | -13.55 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.783300 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 17416213 113 + 22422827 -----UUAGUAUUCGAAUAACAAAAGGUAUCCAGGCUGCACCUGAAUAAAUCCCGCAUUCAGGAUUAAUUUCAGUUCCUGGCUAAAGCUACUUACAAAUUCAGGUAAGCCUUGACCAC -----....................(((....(((((..((((((((.(((((........))))).......((...((((....))))...))..)))))))).)))))..))).. ( -30.50, z-score = -3.26, R) >droSim1.chrX 3542225 111 - 17042790 ----GGUAGUAUUCGUAUAACAAAAGGUAUU---GCUGCAUCUGGAUAAAUCCUGCAUUCAGGAUUAAUUUCAGUUUCUGGCUAAAGCUACUUACCAAUUCUGGUAAGCCCUGGCCAC ----.((((((...((((........)))))---)))))..(((((..(((((((....)))))))...)))))....((((((..(((...(((((....))))))))..)))))). ( -32.50, z-score = -2.67, R) >droSec1.super_17 1261673 102 + 1527944 -------------CGUAUAACAAAAGGUAUU---GCUGCAUCUGGAUAAAUCCUGCAUUCAGGAUUAAUUUCAGUUUCUGGCUAAAGCUACUUACAAAUUCUGGUAAGCCCUGGCCAC -------------.(((..((.....))..)---)).....(((((..(((((((....)))))))...)))))....((((((..((((((..........))).)))..)))))). ( -23.60, z-score = -0.92, R) >droYak2.chrX 16044608 101 + 21770863 GGUAAAAGUCGCACAAAUAUCAAGAGGUACU---GCUGCACC-----AAA----GGAUUUGAAAGUAC-----AUUUCUGGCUAAAGCUACUUACAAACUCUGGCAAAUCCUCACCAU (((.......(((((..((((....)))).)---).)))...-----..(----(((((((..((...-----.....((((....))))..........))..)))))))).))).. ( -20.97, z-score = -1.06, R) >consensus ______UAGUAUUCGAAUAACAAAAGGUAUU___GCUGCACCUGGAUAAAUCCUGCAUUCAGGAUUAAUUUCAGUUUCUGGCUAAAGCUACUUACAAAUUCUGGUAAGCCCUGACCAC .........................(((......(((((..(((((..(((((((....)))))))...)))))....((((....)))).............)).)))....))).. (-12.24 = -13.55 + 1.31)



| Location | 17,416,213 – 17,416,326 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 71.72 |
| Shannon entropy | 0.45825 |
| G+C content | 0.40288 |
| Mean single sequence MFE | -27.62 |
| Consensus MFE | -14.35 |
| Energy contribution | -15.10 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.623634 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 17416213 113 - 22422827 GUGGUCAAGGCUUACCUGAAUUUGUAAGUAGCUUUAGCCAGGAACUGAAAUUAAUCCUGAAUGCGGGAUUUAUUCAGGUGCAGCCUGGAUACCUUUUGUUAUUCGAAUACUAA----- (((.((.((((((((((((((..((..(((((....))(((((...........)))))..)))...))..))))))))).)))))(((((.(....).))))))).)))...----- ( -33.20, z-score = -2.26, R) >droSim1.chrX 3542225 111 + 17042790 GUGGCCAGGGCUUACCAGAAUUGGUAAGUAGCUUUAGCCAGAAACUGAAAUUAAUCCUGAAUGCAGGAUUUAUCCAGAUGCAGC---AAUACCUUUUGUUAUACGAAUACUACC---- .((((.((((((((((......))))...)))))).))))....(((.....(((((((....)))))))....)))....(((---((......)))))..............---- ( -28.70, z-score = -1.89, R) >droSec1.super_17 1261673 102 - 1527944 GUGGCCAGGGCUUACCAGAAUUUGUAAGUAGCUUUAGCCAGAAACUGAAAUUAAUCCUGAAUGCAGGAUUUAUCCAGAUGCAGC---AAUACCUUUUGUUAUACG------------- .((((.(((((((((........)))...)))))).))))....(((.....(((((((....)))))))....)))....(((---((......))))).....------------- ( -24.20, z-score = -1.07, R) >droYak2.chrX 16044608 101 - 21770863 AUGGUGAGGAUUUGCCAGAGUUUGUAAGUAGCUUUAGCCAGAAAU-----GUACUUUCAAAUCC----UUU-----GGUGCAGC---AGUACCUCUUGAUAUUUGUGCGACUUUUACC ..(((((((..(((((((((((((.(((((((....))((....)-----)))))).)))))..----...-----(((((...---.)))))........)))).)))).))))))) ( -24.40, z-score = -0.85, R) >consensus GUGGCCAGGGCUUACCAGAAUUUGUAAGUAGCUUUAGCCAGAAACUGAAAUUAAUCCUGAAUGCAGGAUUUAUCCAGAUGCAGC___AAUACCUUUUGUUAUACGAAUACUA______ .((((.(((((((((........)))...)))))).))))....(((.....(((((((....)))))))....)))......................................... (-14.35 = -15.10 + 0.75)

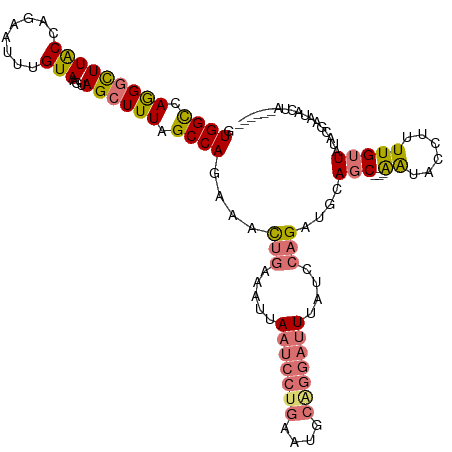

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:53:06 2011