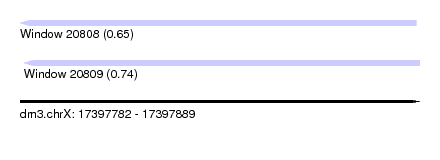
| Sequence ID | dm3.chrX |
|---|---|
| Location | 17,397,782 – 17,397,889 |
| Length | 107 |
| Max. P | 0.737655 |

| Location | 17,397,782 – 17,397,888 |
|---|---|
| Length | 106 |
| Sequences | 7 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 71.71 |
| Shannon entropy | 0.50041 |
| G+C content | 0.44833 |
| Mean single sequence MFE | -28.14 |
| Consensus MFE | -16.60 |
| Energy contribution | -14.37 |
| Covariance contribution | -2.23 |
| Combinations/Pair | 1.52 |
| Mean z-score | -1.06 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.650071 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
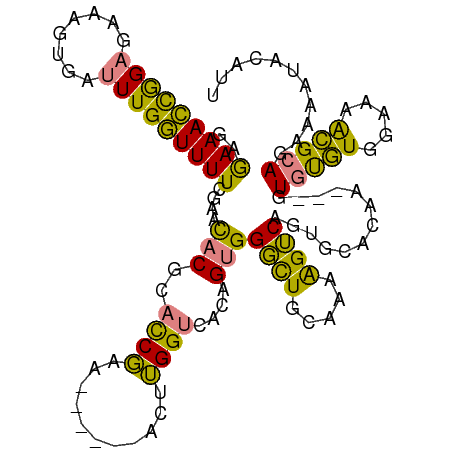
>dm3.chrX 17397782 106 - 22422827 GAGAAACCGGAGAAAGUGAUUUGGUUUUCGAACACGCACCGAA------ACUUGGUCACAGUGGGCUGCAAAAGUCAGUGCACAA---GUGUGUGGAAAACGCAGAAAGUACAUU ...............(((.((((((((((..(((((((((((.------..)))))....(((.(((((....).)))).)))..---)))))).)))))).))))...)))... ( -30.30, z-score = -1.40, R) >droEre2.scaffold_4690 7737050 109 - 18748788 GAGAAACCGGAGAAAGUGAUUUGGUUUUCUAACACGCACCGAA------ACUUGGUCACAGUGGGCUGCAAAAGUCAAUGCACAAGUAGUGUGUGGAAAACGCAUAAAAUACAUU .((((((((((........))))).)))))..(((((((....------(((((....((...((((.....))))..))..))))).))))))).................... ( -26.20, z-score = -0.75, R) >droYak2.chrX 16025973 106 - 21770863 GAGAAACCGGAGAAAGUGAUUUGGUUUUCUAACACGCACCGUA------ACUUGGUCACAGUGGGCUGCAAAAGUCAGUGGACAA---GUGUGUGGAAAACGCAGAAAAUACAUU ...............(((.((((((((((((.(((((((((..------...))))(((....((((.....)))).))).....---))))))))))))).))))...)))... ( -26.50, z-score = -0.61, R) >droSec1.super_17 1243586 106 - 1527944 GAGAAACCGGAGAAAGUGAUUUGGUUUUCGAACACGCACCGAA------ACUUGGUCACAGUGGGCUGCAAAAGUCAGUGCACAA---GUGUGUGGAAAACGCAGAAAGUACAUU ...............(((.((((((((((..(((((((((((.------..)))))....(((.(((((....).)))).)))..---)))))).)))))).))))...)))... ( -30.30, z-score = -1.40, R) >droSim1.chrX 3524003 106 + 17042790 GAGAAACCGGAGAAAGUGAUUUGGUUUUCGAACACGCACCGAA------ACUUGGUCACAGUGGGCUGCAAAAGUCAGUGCACAA---GUGUGUGGAAAACGCAGAAAGUACAUU ...............(((.((((((((((..(((((((((((.------..)))))....(((.(((((....).)))).)))..---)))))).)))))).))))...)))... ( -30.30, z-score = -1.40, R) >dp4.chrXL_group1a 8907872 113 - 9151740 AAAAAAUUAGAAAAGGAAAGUUGAUUUUCACUUGCAAAUCCAAGGGCGGGGCGGGGCGGGGAGGUUUCUUGGGGGCAAUGCGGAA--AAUGCCUAGAAUGGGAAAAUCAGCAAUU ...................((((((((((.(((((...(((......))).....))))).....((((...(((((.(......--).)))))))))...)))))))))).... ( -27.80, z-score = -0.97, R) >droPer1.super_15 1302043 108 - 2181545 AAAAAAUUAGAAAAGGAAAGUUGAUUUUCACUUGCAAACCCAA-----GGGAGGGGCGGGGAGGUUUCUUUGGGGCAAUGCGGAA--AAUGCCUAGAAUGGGAAAAUCAGGAAUU ....................(((((((((..(((((..((((.-----..(((..((......))..)))))))....)))))..--....((......)))))))))))..... ( -25.60, z-score = -0.94, R) >consensus GAGAAACCGGAGAAAGUGAUUUGGUUUUCGAACACGCACCGAA______ACUUGGUCACAGUGGGCUGCAAAAGUCAGUGCACAA___GUGUGUGGAAAACGCAGAAAAUACAUU ..(((((((((........)))))))))....(((..((((...........))))....)))((((.....)))).............(((((.....)))))........... (-16.60 = -14.37 + -2.23)


| Location | 17,397,783 – 17,397,889 |
|---|---|
| Length | 106 |
| Sequences | 7 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 71.01 |
| Shannon entropy | 0.51306 |
| G+C content | 0.45694 |
| Mean single sequence MFE | -28.14 |
| Consensus MFE | -16.60 |
| Energy contribution | -14.37 |
| Covariance contribution | -2.23 |
| Combinations/Pair | 1.52 |
| Mean z-score | -1.10 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.737655 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
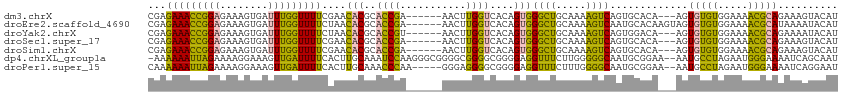
>dm3.chrX 17397783 106 - 22422827 CGAGAAACCGGAGAAAGUGAUUUGGUUUUCGAACACGCACCGA------AACUUGGUCACAGUGGGCUGCAAAAGUCAGUGCACA---AGUGUGUGGAAAACGCAGAAAGUACAU ................(((.((((((((((..(((((((((((------...)))))....(((.(((((....).)))).))).---.)))))).)))))).))))...))).. ( -30.30, z-score = -1.37, R) >droEre2.scaffold_4690 7737051 109 - 18748788 CGAGAAACCGGAGAAAGUGAUUUGGUUUUCUAACACGCACCGA------AACUUGGUCACAGUGGGCUGCAAAAGUCAAUGCACAAGUAGUGUGUGGAAAACGCAUAAAAUACAU ..((((((((((........))))).)))))..(((((((...------.(((((....((...((((.....))))..))..))))).)))))))................... ( -26.20, z-score = -0.72, R) >droYak2.chrX 16025974 106 - 21770863 CGAGAAACCGGAGAAAGUGAUUUGGUUUUCUAACACGCACCGU------AACUUGGUCACAGUGGGCUGCAAAAGUCAGUGGACA---AGUGUGUGGAAAACGCAGAAAAUACAU ................(((.((((((((((((.(((((((((.------....))))(((....((((.....)))).)))....---.))))))))))))).))))...))).. ( -26.50, z-score = -0.59, R) >droSec1.super_17 1243587 106 - 1527944 CGAGAAACCGGAGAAAGUGAUUUGGUUUUCGAACACGCACCGA------AACUUGGUCACAGUGGGCUGCAAAAGUCAGUGCACA---AGUGUGUGGAAAACGCAGAAAGUACAU ................(((.((((((((((..(((((((((((------...)))))....(((.(((((....).)))).))).---.)))))).)))))).))))...))).. ( -30.30, z-score = -1.37, R) >droSim1.chrX 3524004 106 + 17042790 CGAGAAACCGGAGAAAGUGAUUUGGUUUUCGAACACGCACCGA------AACUUGGUCACAGUGGGCUGCAAAAGUCAGUGCACA---AGUGUGUGGAAAACGCAGAAAGUACAU ................(((.((((((((((..(((((((((((------...)))))....(((.(((((....).)))).))).---.)))))).)))))).))))...))).. ( -30.30, z-score = -1.37, R) >dp4.chrXL_group1a 8907873 112 - 9151740 -AAAAAAUUAGAAAAGGAAAGUUGAUUUUCACUUGCAAAUCCAAGGGCGGGGCGGGGCGGGGAGGUUUCUUGGGGGCAAUGCGGAA--AAUGCCUAGAAUGGGAAAAUCAGCAAU -...................((((((((((.(((((...(((......))).....))))).....((((...(((((.(......--).)))))))))...))))))))))... ( -27.80, z-score = -1.14, R) >droPer1.super_15 1302044 108 - 2181545 CAAAAAAUUAGAAAAGGAAAGUUGAUUUUCACUUGCAAACCCAA-----GGGAGGGGCGGGGAGGUUUCUUUGGGGCAAUGCGGAA--AAUGCCUAGAAUGGGAAAAUCAGGAAU .....................(((((((((..(((((..((((.-----..(((..((......))..)))))))....)))))..--....((......))))))))))).... ( -25.60, z-score = -1.11, R) >consensus CGAGAAACCGGAGAAAGUGAUUUGGUUUUCGAACACGCACCGA______AACUUGGUCACAGUGGGCUGCAAAAGUCAGUGCACA___AGUGUGUGGAAAACGCAGAAAAUACAU ...(((((((((........)))))))))....(((..((((...........))))....)))((((.....)))).............(((((.....))))).......... (-16.60 = -14.37 + -2.23)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:53:01 2011