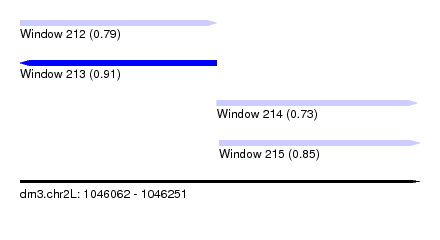
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 1,046,062 – 1,046,251 |
| Length | 189 |
| Max. P | 0.913296 |

| Location | 1,046,062 – 1,046,155 |
|---|---|
| Length | 93 |
| Sequences | 3 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 95.70 |
| Shannon entropy | 0.05924 |
| G+C content | 0.45161 |
| Mean single sequence MFE | -27.77 |
| Consensus MFE | -23.03 |
| Energy contribution | -23.70 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.83 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.794678 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 1046062 93 + 23011544 UCCGUGAAUGGGUCACCAUAUCGCCCUGACAUUAUUAUGCCAGCUUUCCGCUGGUCUAUUUGUGGGUGUGACAUGACCAUUCAAGCUUUUUGU ....(((((((.(((......(((((..(((....((.((((((.....)))))).))..)))))))).....)))))))))).......... ( -29.50, z-score = -2.41, R) >droSim1.chr2L 1016665 93 + 22036055 UCCGCGAAUGGGUCACCAUAUCCCCCUGACAUUAUUAUGCCAGCUUUCUGCUGGUCUAUUUGUGGGUGCGACAUGACCAUUAAAUCUUUUUGU (((((((((((((((...........))))........((((((.....)))))))))))))))))........................... ( -25.00, z-score = -1.64, R) >droSec1.super_14 999478 93 + 2068291 UCCGUGAAUGGGUCACCAUAUCCCCCUGACAUUAUUAUGCCAGCUUUCUGCUGGUCUAUUUGUGGGUGUGACAUGACCAUUCAAUCUUUUUGU ....(((((((.(((.(((((((.....(((....((.((((((.....)))))).))..))))))))))...)))))))))).......... ( -28.80, z-score = -2.73, R) >consensus UCCGUGAAUGGGUCACCAUAUCCCCCUGACAUUAUUAUGCCAGCUUUCUGCUGGUCUAUUUGUGGGUGUGACAUGACCAUUCAAUCUUUUUGU ....(((((((.....(((.((.((((.(((....((.((((((.....)))))).))..)))))).).)).))).))))))).......... (-23.03 = -23.70 + 0.67)


| Location | 1,046,062 – 1,046,155 |
|---|---|
| Length | 93 |
| Sequences | 3 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 95.70 |
| Shannon entropy | 0.05924 |
| G+C content | 0.45161 |
| Mean single sequence MFE | -25.50 |
| Consensus MFE | -25.22 |
| Energy contribution | -25.33 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.99 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.913296 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 1046062 93 - 23011544 ACAAAAAGCUUGAAUGGUCAUGUCACACCCACAAAUAGACCAGCGGAAAGCUGGCAUAAUAAUGUCAGGGCGAUAUGGUGACCCAUUCACGGA ........(.((((((((((((((...((((((..((..(((((.....)))))..))....)))..))).)))))))....))))))).).. ( -26.40, z-score = -1.71, R) >droSim1.chr2L 1016665 93 - 22036055 ACAAAAAGAUUUAAUGGUCAUGUCGCACCCACAAAUAGACCAGCAGAAAGCUGGCAUAAUAAUGUCAGGGGGAUAUGGUGACCCAUUCGCGGA .......((((....))))...((((.((((((..((..(((((.....)))))..))....)))..)))((((..(....)..)))))))). ( -22.40, z-score = -0.49, R) >droSec1.super_14 999478 93 - 2068291 ACAAAAAGAUUGAAUGGUCAUGUCACACCCACAAAUAGACCAGCAGAAAGCUGGCAUAAUAAUGUCAGGGGGAUAUGGUGACCCAUUCACGGA ..........((((((((((((((...((((((..((..(((((.....)))))..))....)))..))).)))))))....))))))).... ( -27.70, z-score = -2.38, R) >consensus ACAAAAAGAUUGAAUGGUCAUGUCACACCCACAAAUAGACCAGCAGAAAGCUGGCAUAAUAAUGUCAGGGGGAUAUGGUGACCCAUUCACGGA ..........((((((((((((((...((((((..((..(((((.....)))))..))....)))..))).)))))))....))))))).... (-25.22 = -25.33 + 0.11)


| Location | 1,046,155 – 1,046,250 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 75.05 |
| Shannon entropy | 0.40047 |
| G+C content | 0.43602 |
| Mean single sequence MFE | -24.16 |
| Consensus MFE | -16.38 |
| Energy contribution | -17.34 |
| Covariance contribution | 0.96 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.727765 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 1046155 95 + 23011544 CUCAGGUCAACGAGGUGCCUCGAAUUGUUAUUCAGUUGACC---------------AGACCU---UAACUGCAGGCAAAUAAAUCAUGUUUGCAAAUGCCCAUUUAUGGGAAG ....((((((((((....)))((((....)))).)))))))---------------......---....((((((((.........))))))))....((((....))))... ( -30.00, z-score = -3.10, R) >droSim1.chr2L 1016758 91 + 22036055 CUCAGGCCAGCGAGGUGCCUCGAAUUGGGACUCAGUUGACC---------------AGACCU---UAUCUGCAGUCAAAUAAAUCAUGAUUGCAAUUGCCCAUUUCUGU---- ....(((((((..(((.((.......)).)))..))))...---------------......---....((((((((.........))))))))...))).........---- ( -23.00, z-score = -0.37, R) >droSec1.super_14 999571 91 + 2068291 CUCAGGUCAACGAGGUGCCUCGAAUUGGAACUCAGUUGACC---------------AGACCU---UAUCUGCAGUCAAAUAAAUCAUGAUUGCAAUUGCCCAUUUCUGU---- ....((((((((((...((.......))..))).)))))))---------------......---....((((((((.........))))))))...............---- ( -25.80, z-score = -2.21, R) >droEre2.scaffold_4929 1087964 89 + 26641161 CCCAGGUAAACGAGGUGCCUCGAAUUAUAAUUCAGUUGGC---------------AAGAUAUGUGUAUCUGCGGUCAAAUAUCUCAUGCUCGCAAUUGCCCAAU--------- ....(((((.((((....))))............((.(((---------------(((((((.((.((.....)))).))))))..)))).))..)))))....--------- ( -21.50, z-score = -1.01, R) >droYak2.chr2L 1021877 113 + 22324452 CUCAGCUUAACGAGGUGCCUCGAAUUAUAAUUCAGUUGGCCAGACCAUAAUAGCCAAGAUAUGUUUACCUGCAGUCAAAUAUAUCAUGUUCGCAAUUGCCCAAUUACGUGAAA ..........((((....)))).............(((((.(........).)))))(((((((((..(....)..)))))))))...(((((((((....))))..))))). ( -20.50, z-score = -0.06, R) >consensus CUCAGGUCAACGAGGUGCCUCGAAUUGUAAUUCAGUUGACC_______________AGACCU___UAUCUGCAGUCAAAUAAAUCAUGAUUGCAAUUGCCCAUUUAUGU____ ....((((((((((....)))((((....)))).)))))))............................((((((((.........))))))))................... (-16.38 = -17.34 + 0.96)

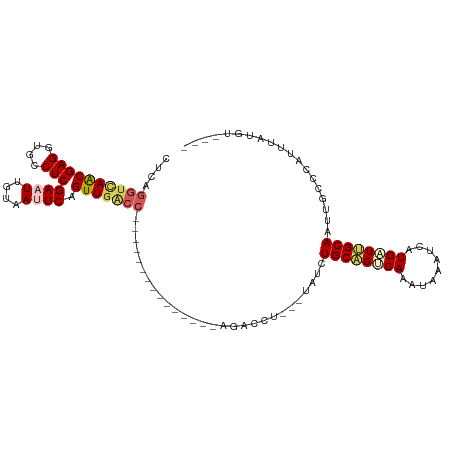
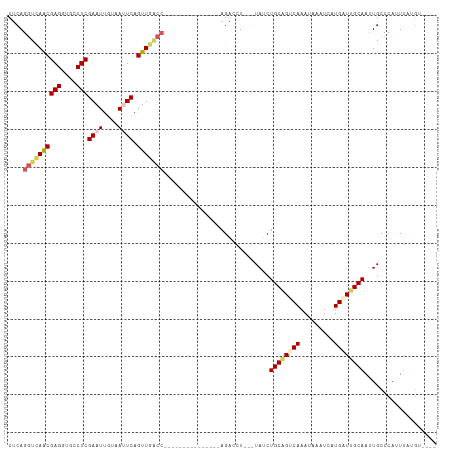
| Location | 1,046,156 – 1,046,251 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 74.39 |
| Shannon entropy | 0.40906 |
| G+C content | 0.43233 |
| Mean single sequence MFE | -24.72 |
| Consensus MFE | -16.38 |
| Energy contribution | -17.34 |
| Covariance contribution | 0.96 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.847341 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 1046156 95 + 23011544 UCAGGUCAACGAGGUGCCUCGAAUUGUUAUUCAGUUGACC---------------AGACCU---UAACUGCAGGCAAAUAAAUCAUGUUUGCAAAUGCCCAUUUAUGGGAAGG ...((((((((((....)))((((....)))).)))))))---------------...(((---(...((((((((.........))))))))....((((....)))))))) ( -32.80, z-score = -3.73, R) >droSim1.chr2L 1016759 90 + 22036055 UCAGGCCAGCGAGGUGCCUCGAAUUGGGACUCAGUUGACC---------------AGACCU---UAUCUGCAGUCAAAUAAAUCAUGAUUGCAAUUGCCCAUUUCUGU----- ...(((((((..(((.((.......)).)))..))))...---------------......---....((((((((.........))))))))...))).........----- ( -23.00, z-score = -0.48, R) >droSec1.super_14 999572 90 + 2068291 UCAGGUCAACGAGGUGCCUCGAAUUGGAACUCAGUUGACC---------------AGACCU---UAUCUGCAGUCAAAUAAAUCAUGAUUGCAAUUGCCCAUUUCUGU----- ...((((((((((...((.......))..))).)))))))---------------......---....((((((((.........))))))))...............----- ( -25.80, z-score = -2.30, R) >droEre2.scaffold_4929 1087965 88 + 26641161 CCAGGUAAACGAGGUGCCUCGAAUUAUAAUUCAGUUGGC---------------AAGAUAUGUGUAUCUGCGGUCAAAUAUCUCAUGCUCGCAAUUGCCCAAU---------- ...(((((.((((....))))............((.(((---------------(((((((.((.((.....)))).))))))..)))).))..)))))....---------- ( -21.50, z-score = -1.07, R) >droYak2.chr2L 1021878 113 + 22324452 UCAGCUUAACGAGGUGCCUCGAAUUAUAAUUCAGUUGGCCAGACCAUAAUAGCCAAGAUAUGUUUACCUGCAGUCAAAUAUAUCAUGUUCGCAAUUGCCCAAUUACGUGAAAG .........((((....)))).............(((((.(........).)))))(((((((((..(....)..)))))))))...(((((((((....))))..))))).. ( -20.50, z-score = -0.27, R) >consensus UCAGGUCAACGAGGUGCCUCGAAUUGUAAUUCAGUUGACC_______________AGACCU___UAUCUGCAGUCAAAUAAAUCAUGAUUGCAAUUGCCCAUUUAUGU_____ ...((((((((((....)))((((....)))).)))))))............................((((((((.........)))))))).................... (-16.38 = -17.34 + 0.96)

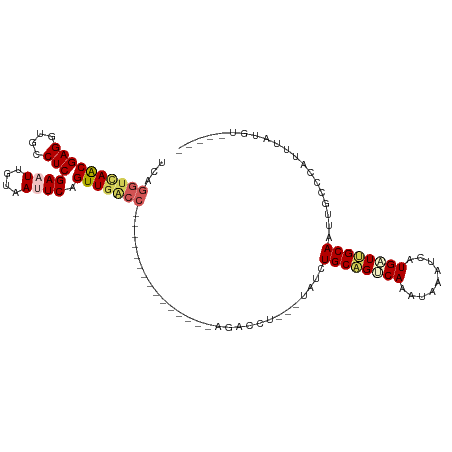
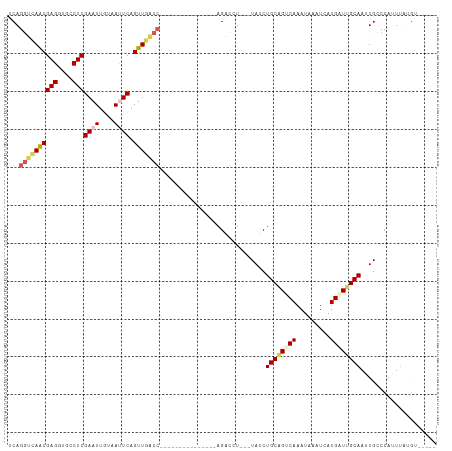
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:07:38 2011