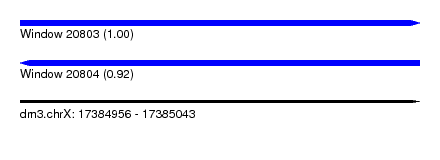
| Sequence ID | dm3.chrX |
|---|---|
| Location | 17,384,956 – 17,385,043 |
| Length | 87 |
| Max. P | 0.998374 |

| Location | 17,384,956 – 17,385,043 |
|---|---|
| Length | 87 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 76.45 |
| Shannon entropy | 0.40887 |
| G+C content | 0.47620 |
| Mean single sequence MFE | -30.98 |
| Consensus MFE | -25.43 |
| Energy contribution | -26.18 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.91 |
| Structure conservation index | 0.82 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.34 |
| SVM RNA-class probability | 0.998374 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
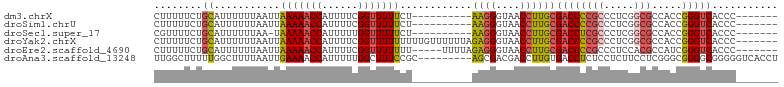
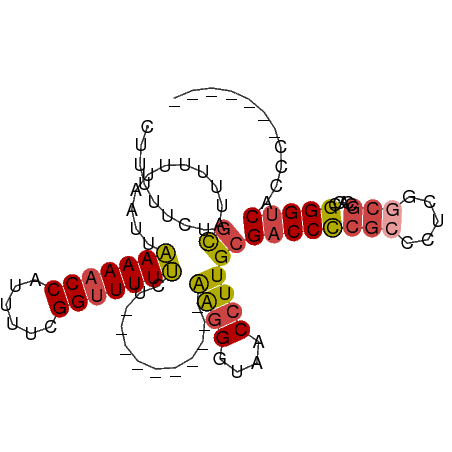
>dm3.chrX 17384956 87 + 22422827 -------GGGUGACCCGGUGGCGCCGAGGGCGGGGUCGCAAGGUUACCCUU----------AGAAAAACCGAAAAUGGUUUUUAAUUAAAAAAUGCAGAAAAAG -------((((((((..(((((.(((....))).)))))..))))))))..----------..(((((((......)))))))..................... ( -34.00, z-score = -3.83, R) >droSim1.chrU 760955 87 - 15797150 -------GGGUGACCCGGUGGCGCCGAGGGCGGGGUCGCAAGGUUACCCUU----------AGAAAAACCGAAAAUGGUUUUUAAUUAAAAAAUGCAGAAAAAG -------((((((((..(((((.(((....))).)))))..))))))))..----------..(((((((......)))))))..................... ( -34.00, z-score = -3.83, R) >droSec1.super_17 1230664 86 + 1527944 -------GGGUGACCCGGUGGCGCCGAGGGCGAGGUCGCAAGGUUACCCUU----------AGAAAAACCAAAAAUGGUUUUUA-UUAAAAAAUGCAGAAAACG -------((((((((..((((((((...)))...)))))..))))))))((----------((((((((((....)))))))).-))))............... ( -29.90, z-score = -2.87, R) >droYak2.chrX 16012793 97 + 21770863 -------GGGUGACCCGGUGGCGCCGAGGGCGGGGUCGCAAGGUUACCCUCUAAAAAACAAAAAAAAACCGAAAAUGGUUUUUAAUUAAAAAAUGCAGAAAAAG -------((((((((..(((((.(((....))).)))))..))))))))..............(((((((......)))))))..................... ( -33.90, z-score = -3.71, R) >droEre2.scaffold_4690 7724247 92 + 18748788 -------GGGUGACCCGAUGGCGUGGAGGGCGGGGUCGCAAGGUUACCCUCUAAAA-----AAAAAAACCGAAAAUGGUUUUUAAUUAAAAAAUGCAGAAAAAG -------((....)).....((((((((((...(..(....)..).))))))....-----..(((((((......))))))).........))))........ ( -26.00, z-score = -2.04, R) >droAna3.scaffold_13248 1785646 95 - 4840945 AGGUGACCCCCGCCCCGCCCGAGGAAGAGGAGAGGUCACAAGGUCGUCGCU---------GCGGAAAGCCAAAAAUGGUUUUCAAUUAAAAGCCAAAAAGCCAA ..(((((((((...((......))....)).).))))))..(((....(((---------...((((((((....)))))))).......)))......))).. ( -28.10, z-score = -1.18, R) >consensus _______GGGUGACCCGGUGGCGCCGAGGGCGGGGUCGCAAGGUUACCCUU__________AGAAAAACCGAAAAUGGUUUUUAAUUAAAAAAUGCAGAAAAAG .......((((((((..(((((.(((....))).)))))..))))))))..............((((((((....))))))))..................... (-25.43 = -26.18 + 0.76)


| Location | 17,384,956 – 17,385,043 |
|---|---|
| Length | 87 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 76.45 |
| Shannon entropy | 0.40887 |
| G+C content | 0.47620 |
| Mean single sequence MFE | -23.85 |
| Consensus MFE | -16.41 |
| Energy contribution | -16.75 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.917121 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 17384956 87 - 22422827 CUUUUUCUGCAUUUUUUAAUUAAAAACCAUUUUCGGUUUUUCU----------AAGGGUAACCUUGCGACCCCGCCCUCGGCGCCACCGGGUCACCC------- ........((...........(((((((......)))))))..----------((((....))))))(((((((((...)))).....)))))....------- ( -23.30, z-score = -2.08, R) >droSim1.chrU 760955 87 + 15797150 CUUUUUCUGCAUUUUUUAAUUAAAAACCAUUUUCGGUUUUUCU----------AAGGGUAACCUUGCGACCCCGCCCUCGGCGCCACCGGGUCACCC------- ........((...........(((((((......)))))))..----------((((....))))))(((((((((...)))).....)))))....------- ( -23.30, z-score = -2.08, R) >droSec1.super_17 1230664 86 - 1527944 CGUUUUCUGCAUUUUUUAA-UAAAAACCAUUUUUGGUUUUUCU----------AAGGGUAACCUUGCGACCUCGCCCUCGGCGCCACCGGGUCACCC------- ........((.........-.((((((((....))))))))..----------((((....))))))(((((((((...)))).....)))))....------- ( -23.10, z-score = -1.86, R) >droYak2.chrX 16012793 97 - 21770863 CUUUUUCUGCAUUUUUUAAUUAAAAACCAUUUUCGGUUUUUUUUUGUUUUUUAGAGGGUAACCUUGCGACCCCGCCCUCGGCGCCACCGGGUCACCC------- ........((......(((..(((((((......)))))))..))).......((((....))))))(((((((((...)))).....)))))....------- ( -25.40, z-score = -2.28, R) >droEre2.scaffold_4690 7724247 92 - 18748788 CUUUUUCUGCAUUUUUUAAUUAAAAACCAUUUUCGGUUUUUUU-----UUUUAGAGGGUAACCUUGCGACCCCGCCCUCCACGCCAUCGGGUCACCC------- ........((.......((..(((((((......)))))))..-----))...((((....))))))(((((.((.......))....)))))....------- ( -19.60, z-score = -1.67, R) >droAna3.scaffold_13248 1785646 95 + 4840945 UUGGCUUUUUGGCUUUUAAUUGAAAACCAUUUUUGGCUUUCCGC---------AGCGACGACCUUGUGACCUCUCCUCUUCCUCGGGCGGGGCGGGGGUCACCU ..((((....)))).......((((.(((....))).))))...---------............(((((((((((((.((....)).)))).))))))))).. ( -28.40, z-score = -0.03, R) >consensus CUUUUUCUGCAUUUUUUAAUUAAAAACCAUUUUCGGUUUUUCU__________AAGGGUAACCUUGCGACCCCGCCCUCGGCGCCACCGGGUCACCC_______ ........((...........(((((((......)))))))............((((....))))))((((((((.....))).....)))))........... (-16.41 = -16.75 + 0.34)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:52:57 2011