| Sequence ID | dm3.chrX |
|---|---|
| Location | 17,373,832 – 17,373,908 |
| Length | 76 |
| Max. P | 0.964597 |

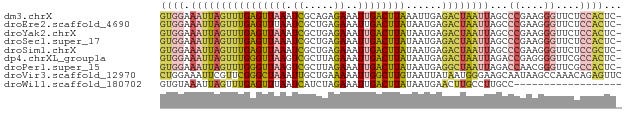
| Location | 17,373,832 – 17,373,908 |
|---|---|
| Length | 76 |
| Sequences | 9 |
| Columns | 77 |
| Reading direction | reverse |
| Mean pairwise identity | 78.79 |
| Shannon entropy | 0.47769 |
| G+C content | 0.37984 |
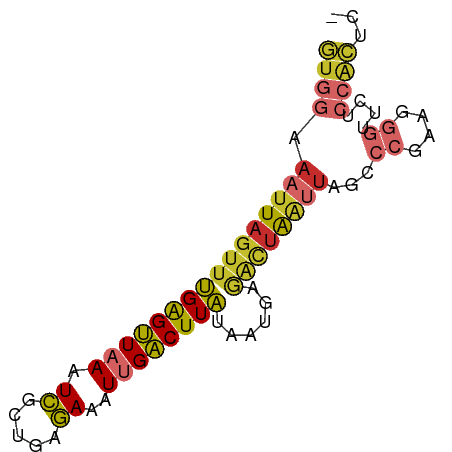
| Mean single sequence MFE | -18.44 |
| Consensus MFE | -10.02 |
| Energy contribution | -10.58 |
| Covariance contribution | 0.55 |
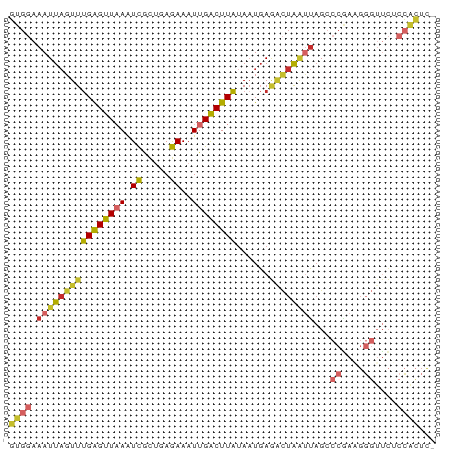
| Combinations/Pair | 1.46 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.74 |
| SVM RNA-class probability | 0.964597 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 17373832 76 - 22422827 GUGGAAAUUAGUUUGAGUUAAAUCGCAGAGAAAUUGACUUAAAUUGAGACUAAUUAGCCCGAAGGGUUCUCCACUC- (((((..((((((((((((((.((.....))..))))))))))))))........(((((...))))).)))))..- ( -22.40, z-score = -3.35, R) >droEre2.scaffold_4690 7713808 76 - 18748788 GUGGAAAUUAGUUUGAGUUUAAUCGCUGAGAAAUUGACUUAUAAUGAGACUAAUUAGCCCGAAGGGUUCUCCACUC- (((((((((((((((((((...((.....))....))))))......))))))))(((((...))))).)))))..- ( -19.20, z-score = -2.14, R) >droYak2.chrX 16001544 76 - 21770863 GUGGAAAUUAGUUUGAGUUAAAUCGCUGAGAAAUUGACUUAUAAUGAGACUAAUUAGCCCGAAGGGUUCUCCACUC- (((((((((((((((((((((.((.....))..))))))))......))))))))(((((...))))).)))))..- ( -20.60, z-score = -2.77, R) >droSec1.super_17 1219694 76 - 1527944 GUGGAAAUUAGUUUGAGUUAAAUCGCUGAGAAAUUGACUUAUAAUGAGACUAAUUAGCCCGAAGGGUUCUCCACUC- (((((((((((((((((((((.((.....))..))))))))......))))))))(((((...))))).)))))..- ( -20.60, z-score = -2.77, R) >droSim1.chrX 3499695 76 + 17042790 GUGGAAAUUAGUUUGAGUUAAAUCGCUGAGAAAUUGACUUAUAAUGAGACUAAUUAGCCCGAAGGGUUCUCCGCUC- (((((((((((((((((((((.((.....))..))))))))......))))))))(((((...))))).)))))..- ( -20.20, z-score = -2.44, R) >dp4.chrXL_group1a 8881181 76 - 9151740 GUGGAAAUUAGUUUGGGUUAAGUCGCUUAGAAAUUGACUUAUAAUGAGACUAAUUAGACCGAGGGGUUCGCCACUC- ((((.(((((((((.(..(((((((.........)))))))...).))))))))).((((....))))..))))..- ( -21.70, z-score = -2.52, R) >droPer1.super_15 1275296 76 - 2181545 GUGGAAAUUAGUUUGGGUUAAGUCGCUUAGAAAUUGACUUAUAAUGAGGCUAAUUAGACCAACGGGUUCGCCACUC- ((((.(((((((((.(..(((((((.........)))))))...).))))))))).((((....))))..))))..- ( -21.00, z-score = -2.35, R) >droVir3.scaffold_12970 6103046 77 + 11907090 CUGGAAAUUCGUUCGGGCUAAAUUGCUGAAAAAUUGGCUUGUAAUUAUAAUGGGAAGCAAUAAGCCAAACAGAGUUC (((........(((((.(......))))))...(((((((((.................))))))))).)))..... ( -15.13, z-score = -0.89, R) >droWil1.scaffold_180702 542727 59 - 4511350 GUGUAAAUUAGUUUGAGUUUAAUCAUCUAGAAAUUGACUUAUAAUGAACUUGCCUUGCC------------------ ..((((...((((((((((...((.....))....))))).....)))))....)))).------------------ ( -5.10, z-score = 0.40, R) >consensus GUGGAAAUUAGUUUGAGUUAAAUCGCUGAGAAAUUGACUUAUAAUGAGACUAAUUAGCCCGAAGGGUUCUCCACUC_ ((((.((((((((((((((((.((.....))..))))))))......))))))))...((....))....))))... (-10.02 = -10.58 + 0.55)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:52:53 2011