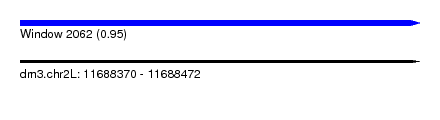
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 11,688,370 – 11,688,472 |
| Length | 102 |
| Max. P | 0.952926 |

| Location | 11,688,370 – 11,688,472 |
|---|---|
| Length | 102 |
| Sequences | 8 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 74.59 |
| Shannon entropy | 0.46517 |
| G+C content | 0.52193 |
| Mean single sequence MFE | -29.81 |
| Consensus MFE | -21.22 |
| Energy contribution | -20.55 |
| Covariance contribution | -0.67 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.952926 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 11688370 102 + 23011544 CCCAAAUGGGCCAACAUGGC----GCUCGUGGCAGUCAAUAAUAAGCAGGCGACAUGUUGGCCAGAGCACACUUUCGGCCAUCUCCCAAAAAAAAGGGUAAGAAAU----- .((.....((((((((((.(----((..((...............))..))).)))))))))).(((......)))))...((((((........)))..)))...----- ( -30.46, z-score = -1.30, R) >droSim1.chr2L 11497884 101 + 22036055 CCCAAAUGGGCCAACAUGGC----GCUCGUGGCAGUCAAUAAUAAGCAGGCGACAUGUUGGCCAGAGCACACUUUCGGCCAUCACCCAAAAAAA-GGGAAAGAAAU----- (((.....((((((((((.(----((..((...............))..))).)))))))))).(.((.(......))))..............-)))........----- ( -30.46, z-score = -1.48, R) >droSec1.super_3 7080396 101 + 7220098 CCCAAAUGGGCCAACAUGGC----GCUCGUGGCAGUCAAUAAUAAGCAGGCGACAUGUUGGCCAGAGCACACUUUCGGCCAUCUCCCAAAAAAA-GGGAAAGAAAU----- .((.....((((((((((.(----((..((...............))..))).)))))))))).(((......))))).....((((.......-)))).......----- ( -31.16, z-score = -1.53, R) >droAna3.scaffold_12916 9406727 107 - 16180835 CCCAAAUGGGCCAACAUGGC----GCUCGUGGCAGUCAAUAAUAAGCAGGCGACAUGUUGGCCAGAGCUCACUUUCGGCCAUCUCCACGGCACGAAGGAAAAAAGUGUCAU ........((((((((((.(----((..((...............))..))).))))))))))........(((((((((........))).))))))............. ( -37.16, z-score = -2.12, R) >dp4.chr4_group3 2552599 84 + 11692001 CCCAAAUGGGCCGACAUGGCAGUCGCUCGUGGCAGUCAAUAAUAAGCAGGCGACAUGUUGGCCAGAGUACACUUUCGGCCCAAC--------------------------- ......((((((((..((((.(((((..((...............))..)))))......))))(((....)))))))))))..--------------------------- ( -29.66, z-score = -1.97, R) >droWil1.scaffold_180772 5152123 86 - 8906247 CCCAAAUGGGCCGGCAUGGC----GCUUGUGGCAGUCAAUAAUAAGCAGGCGACAUGUUGGCCAAAGAAAGUACAUAAAAAGAACCAGAU--------------------- ........((((((((((.(----((((((...............))))))).))))))))))...........................--------------------- ( -27.96, z-score = -2.58, R) >droVir3.scaffold_12963 6260020 77 + 20206255 CCCAAAUGGGCCAACAUGGC----GCUUGUGGCAGUCAAUAAUAAGCAUGCGACAUGUUGGCUGGAGUACACUUU-GGCCCC----------------------------- .......(((((((..((..----((((....(((((((((....(....)....))))))))))))).))..))-))))).----------------------------- ( -25.90, z-score = -0.70, R) >droMoj3.scaffold_6500 25402620 78 + 32352404 CCCAAAUGGGCCAACAUGGC----GCUCGUGGCAGUCAAUAAUAAGCAUGCGACAUGUUGGCUGGAGCACACUUUCGGCCCC----------------------------- .......(((((......(.----((((....(((((((((....(....)....))))))))))))).)......))))).----------------------------- ( -25.70, z-score = -0.55, R) >consensus CCCAAAUGGGCCAACAUGGC____GCUCGUGGCAGUCAAUAAUAAGCAGGCGACAUGUUGGCCAGAGCACACUUUCGGCCAUCUCC__A______________________ ........((((((((((((....))((((.((............))..))))))))))))))................................................ (-21.22 = -20.55 + -0.67)

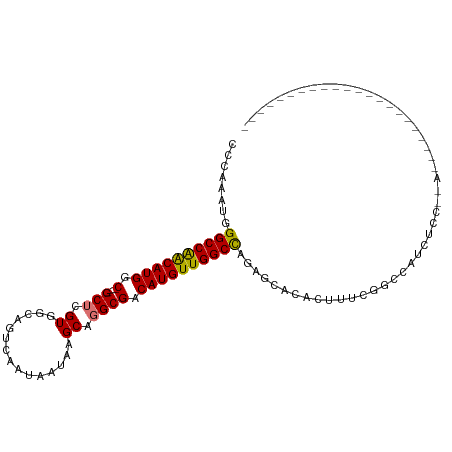
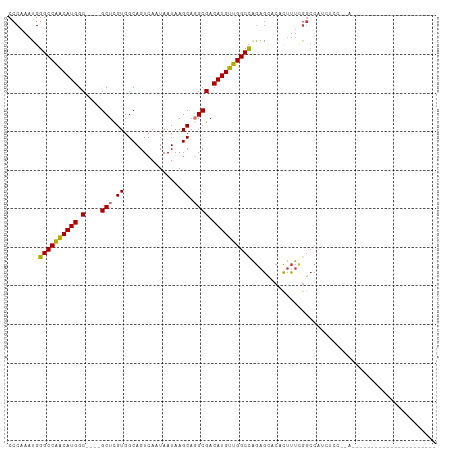
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:33:20 2011