| Sequence ID | dm3.chrX |
|---|---|
| Location | 17,354,398 – 17,354,495 |
| Length | 97 |
| Max. P | 0.548543 |

| Location | 17,354,398 – 17,354,495 |
|---|---|
| Length | 97 |
| Sequences | 13 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 75.53 |
| Shannon entropy | 0.57433 |
| G+C content | 0.55751 |
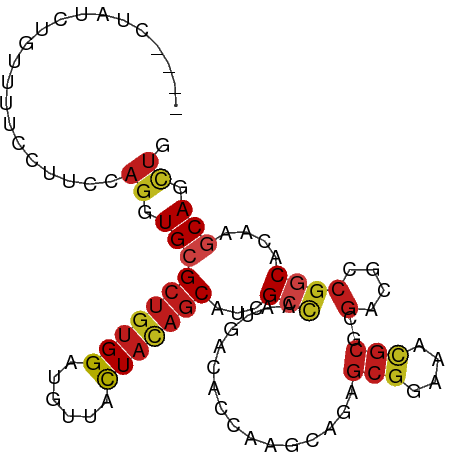
| Mean single sequence MFE | -29.32 |
| Consensus MFE | -19.15 |
| Energy contribution | -19.58 |
| Covariance contribution | 0.43 |
| Combinations/Pair | 1.26 |
| Mean z-score | -0.38 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.548543 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
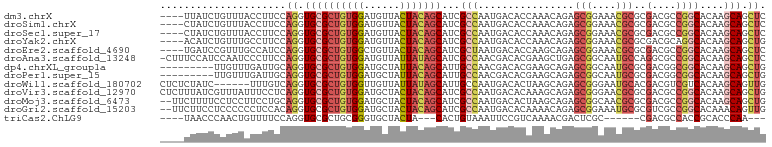
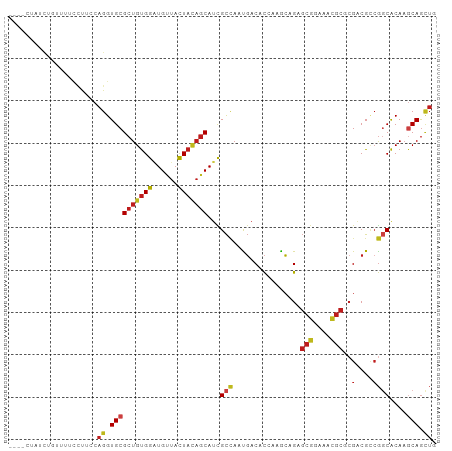
>dm3.chrX 17354398 97 - 22422827 ----UUAUCUGUUUACCUUCCAGGUGCGCUGUGGAUGUUACUACAGCAUCGCCAAUGACACCAAACAGAGCGGAAACGCGCGACGCCGGCACAAGCAGCUC ----....((((((.....((.((((.(((((((......))))))).((((...((....))......(((....)))))))))))))...))))))... ( -31.80, z-score = -1.68, R) >droSim1.chrX 3481750 97 + 17042790 ----CUAUCUGUUUACCUUCCAGGUGCGCUGUGGAUGUUACUACAGCAUCGCCAAUGACACCAAACAGAGCGGAAACGCGCGACGCCGGCACAAGCAGCUC ----....((((((.....((.((((.(((((((......))))))).((((...((....))......(((....)))))))))))))...))))))... ( -31.80, z-score = -1.57, R) >droSec1.super_17 1200032 97 - 1527944 ----CUAUCUGUUUACCUUCCAGGUGCGCUGUGGAUGUUACUACAGCAUCGCCAAUGACACCAAACAGAGCGGAAACGCGCGACGCCGGCACAAGCAGCUC ----....((((((.....((.((((.(((((((......))))))).((((...((....))......(((....)))))))))))))...))))))... ( -31.80, z-score = -1.57, R) >droYak2.chrX 15982302 97 - 21770863 ----ACAUCUGUUUGCCUUCCAGGUGCGCUGUGGAUGUUACUACAGCAUCGCCAAUGACACCAAACAGAGCGGAAACGCGCGACGCAGGCACAAGCAGCUG ----...((((((((...((..((((.(((((((......)))))))..))))...))...))))))))(((....))).....((..((....)).)).. ( -34.40, z-score = -1.62, R) >droEre2.scaffold_4690 7693938 97 - 18748788 ----UGAUCCGUUUGCCAUCCAGGUGCGCUGUGGCUGUUACUACAGCAUCGCUAAUGACACCAAGCAGAGCGGAAACGCGCGACGCCGGCACAAGCAGCUC ----......(((((....((.((((.(((((((......))))))).((((...((....))......(((....)))))))))))))..)))))..... ( -32.60, z-score = -0.24, R) >droAna3.scaffold_13248 1754915 100 + 4840945 -CUUUCCAUCCAAUCCCUUCCAGGUGCGCUGUGGAUGUUAUUAUAGCAUCGCCAACGACACGAAGCUGAGCGGCAAUGCCAGGCGCCGGCACAAGCAGCUC -...............((((...(((((.((..(((((((...)))))))..)).)).)))))))..((((.((..((((.(....)))))...)).)))) ( -26.10, z-score = 1.06, R) >dp4.chrXL_group1a 8856146 92 - 9151740 ---------UUGUUUGAUUGCAGGUGCGCUGUGGAUGCUAUUACAGCAUUGCCAACGACACGAAGCAGAGCGGCAAUGCGCGACGGCGGCACAAGCAGCUG ---------........((((..(((((((((.(.((......))((((((((..((...))..((...)))))))))).).))))).))))..))))... ( -31.10, z-score = 0.17, R) >droPer1.super_15 1248321 92 - 2181545 ---------UUGUUUGAUUGCAGGUGCGCUGUGGAUGCUAUUACAGCAUUGCCAACGACACGAAGCAGAGCGGCAAUGCGCGACGGCGGCACAAGCAGCUG ---------........((((..(((((((((.(.((......))((((((((..((...))..((...)))))))))).).))))).))))..))))... ( -31.10, z-score = 0.17, R) >droWil1.scaffold_180702 523315 95 - 4511350 CUCUCUAUC------UUUGUCAGGUGCGCUGUGGUUGUUAUUAUAGCAUUGCCAAUGACACUAAGCAGAGCGGGAAUGCACGACGUCGUCACAAGCAGUUG .((((...(------(((((..((((((...(((((((((...)))))..)))).)).))))..)))))).)))).((((((....))).....))).... ( -21.50, z-score = 1.42, R) >droVir3.scaffold_12970 6081279 101 + 11907090 CUCUUUAUCGUUUAUUUCCUCAGGUGCGCUGUGGAUGCUACUACAGCAUCGCCAAUGACACAAAGCAGAGCGGGAACGCGCGACGCCGGCACAAGCAGCUG ....................(((.((((((((((......)))))))...(((................(((....)))((...)).)))....))).))) ( -29.60, z-score = 0.16, R) >droMoj3.scaffold_6473 15821081 99 - 16943266 --UUCUUUUCCUCCUUCCUGCAGGUGCGCUGUGGAUGCUACUACAGCAUCGCCAAUGACACUAAGCAGAGCGGCAACGCGCGACGCCGGCACAAGCAGCUG --...............((((..(((((((((((......))))))).((((...((........))..(((....))))))).....))))..))))... ( -33.80, z-score = -0.68, R) >droGri2.scaffold_15203 7516585 99 - 11997470 --UUCUUCCUCCCCCCUCCACAGGUGCGCUGUGGAUGCUACUACAGCAUCGCCAAUGACACAAAACAGAGCGGAAAUGCGCGUCGCCGGCACAAACAGUUG --................(((((.....)))))((((((.....))))))(((..((((.(......).(((......)))))))..)))........... ( -24.80, z-score = 0.16, R) >triCas2.ChLG9 4192214 85 + 15222296 ----UAACCCAACUGUUUUCCAGGUGCGCUGCGGGUGCUACUA---CACUGUAAAUUCCGUCAAAACGACUCGC------CGACGCCACCGCACCCAA--- ----..................((((((..(((((((.(((..---....)))......(((.....))).)))------)..)))...))))))...--- ( -20.70, z-score = -0.71, R) >consensus ____CUAUCUGUUUUCCUUCCAGGUGCGCUGUGGAUGUUACUACAGCAUCGCCAAUGACACCAAGCAGAGCGGAAACGCGCGACGCCGGCACAAGCAGCUG .....................((.((((((((((......)))))))...(((................(((....)))..(....))))....))).)). (-19.15 = -19.58 + 0.43)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:52:49 2011