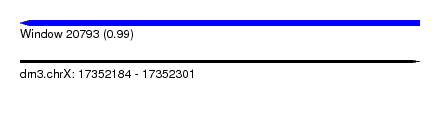
| Sequence ID | dm3.chrX |
|---|---|
| Location | 17,352,184 – 17,352,301 |
| Length | 117 |
| Max. P | 0.991661 |

| Location | 17,352,184 – 17,352,301 |
|---|---|
| Length | 117 |
| Sequences | 13 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 79.98 |
| Shannon entropy | 0.48063 |
| G+C content | 0.57318 |
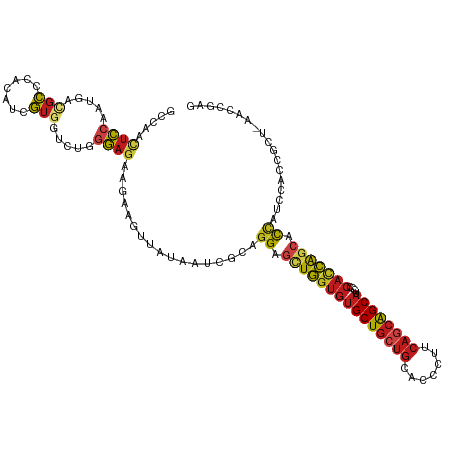
| Mean single sequence MFE | -39.91 |
| Consensus MFE | -26.55 |
| Energy contribution | -27.13 |
| Covariance contribution | 0.58 |
| Combinations/Pair | 1.46 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.49 |
| SVM RNA-class probability | 0.991661 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
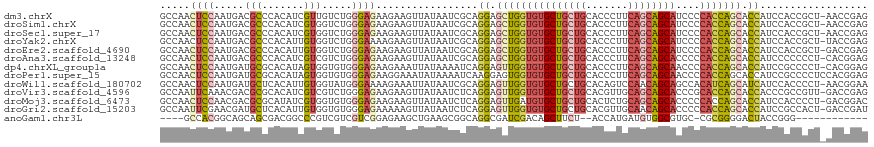
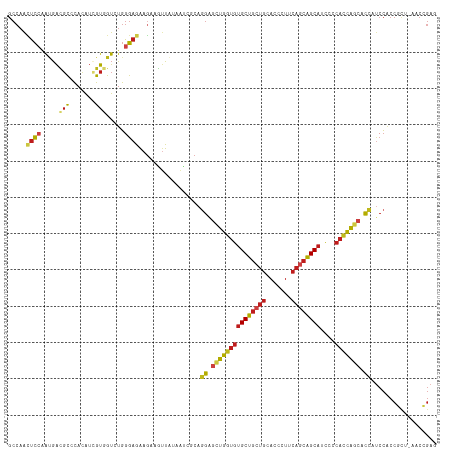
>dm3.chrX 17352184 117 - 22422827 GCCAACUCCAAUGACGCCCACAUCGUUGUCUGGGAGAAGAAGUUAUAAUCGCAGGAGCUGGUGUGCUGCUGCACCCUUCAGCAGCAUCCCCACCAGCACCAUCCACCGCU-AACCGAG ((...((((...((((.(......).))))..)))).................((.(((((((((((((((.......))))))))....))))))).)).......)).-....... ( -38.20, z-score = -2.62, R) >droSim1.chrX 3479536 117 + 17042790 GCCAACUCCAAUGACGCCCACAUCGUGGUCUGGGAGAAGAAGUUAUAAUCGCAGGAGCUGGUGUGCUGCUGCACCCUUCAGCAGCAUCCCCACCAGCACCAUCCACCGCU-AACCGAG ((...((((...((...((((...))))))..)))).................((.(((((((((((((((.......))))))))....))))))).)).......)).-....... ( -38.40, z-score = -2.03, R) >droSec1.super_17 1197818 117 - 1527944 GCCAACUCCAAUGACGCCCACAUCGUGGUCUGGGAGAAGAAGUUAUAAUCGCAGGAGCUGGUGUGCUGCUGCACCCUUCAGCAGCAUCCCCACCAGCACCAUCCACCGCU-AACCGAG ((...((((...((...((((...))))))..)))).................((.(((((((((((((((.......))))))))....))))))).)).......)).-....... ( -38.40, z-score = -2.03, R) >droYak2.chrX 15980079 117 - 21770863 GCCAACUCCAAUGACGCCCACAUUGUGGUCUGGGAAAAGAAGUUAUAAUCGCAGGAGCUGGUGUGCUGCUGCACCCUUCAGCAGCAUCCCCACCAGCACCAUCCACCGCU-UACCGAG ((((((..(((((.......)))))..)).))(((...((........))...((.(((((((((((((((.......))))))))....))))))).)).)))...)).-....... ( -37.30, z-score = -1.93, R) >droEre2.scaffold_4690 7691730 117 - 18748788 GCCAACUCCAAUGACGCCCACAUUGUGGUCUGGGAGAAGAAGUUAUAAUCGCAGGAGCUGGUGUGCUGCUGCACCCUUCAGCAGCAUCCCCACCAGCACCAUCCACCGCU-GACCGAG .....((((((((.......))))).(((((((..((.((........))...((.(((((((((((((((.......))))))))....))))))).)).))..)))..-))))))) ( -40.70, z-score = -2.19, R) >droAna3.scaffold_13248 1752641 117 + 4840945 GCCAACUCCAAUGACGCCCACAUCGUCGUCUGGGAGAAGAAGUUAUAAUCGCAGGAGCUGGUGUGCUGCUGCACCCUUCAGCAGCACCCCCACCAGCACCAUCCCCCCCU-CACGGAG .....((((.((((((.......))))))..((((...((........))...((.(((((((((((((((.......)))))))))....)))))).)).)))).....-...)))) ( -42.70, z-score = -3.82, R) >dp4.chrXL_group1a 8853791 117 - 9151740 GCCAACUCCAAUGAUGCGCACAUAGUGGUGUGGGAGAAGAAAUUAUAAAAUCAGGAGUUGGUGUGCUGCUGCACCCUUCAGCAGCAACCCCACCAGCACCAUCCGCCCCU-CACGGAG .....((((.......(((.....)))(((.(((..((....)).........((.(((((((((((((((.......))))))))....))))))).))......))).-))))))) ( -41.10, z-score = -2.59, R) >droPer1.super_15 1245964 118 - 2181545 GCCAACUCCAAUGAUGCGCACAUAGUGGUGUGGGAGAAGGAAAUAUAAAAUCAAGGAGUGGUGUGCUGCUGCACCCUUCAGCAGCAACCCCACCAGCACCAUCCGCCCCUCCACGGAG .....((((....(((....))).((((.(.(((....(((.((.....))...((.((((((((((((((.......))))))))....))))).).)).))).)))).)))))))) ( -38.10, z-score = -1.29, R) >droWil1.scaffold_180702 520984 117 - 4511350 GCCAACUCCAAUGAUGCUCACAUUGUGGUAUGGGAAAAGAAAUUAUAAUCGCAGGAGUUGGUGUGCUGCUGCACAGUCCAACAGCAGCCACAUCAGCAUCAUCCACCCCU-AACGGAA .(((((..(((((.......)))))..)).)))....................((((((((((((((((((..........)))))).)))))))))....)))...((.-...)).. ( -35.70, z-score = -1.47, R) >droVir3.scaffold_4596 1320 117 - 1902 GCCAAUUCAAACGACGCGCACAUCGUCGUCUGGGAGAAGAAGUUAUAAUCUCAGGAGUUGGUGUGCUGCUGCACGUUGCAGCAGCACCCGCACCAGCACCACCCGCCGUU-GACCGAG ((((((((..((((((.......))))))((((((.............))))))))))))))((((((((((.....)))))))))).((...((((..........)))-)..)).. ( -44.12, z-score = -2.64, R) >droMoj3.scaffold_6473 15818724 117 - 16943266 GCCAACUCCAACGACGCGCAUAUCGUGGUGUGGGAGAAGAAGUUAUAAUCUCAGGAGUUGAUGUGCUGCUGCACUCUGCAGCAGCACCCCCACCAGCACCAUCCACCCCU-GACGGAC (((((((((..(.((((.(((...))))))).)((((...........)))).)))))))..((((((((((.....))))))))))........)).((.((.......-)).)).. ( -42.20, z-score = -2.53, R) >droGri2.scaffold_15203 7514248 117 - 11997470 GCCAAUUCGAACGAUGCUCACAUUGUGGUGUGGGAGAAAAAGUUAUAAUCUCAGGAGUUGGUGUGCUGCUGCACGUUGCAACAGCACCCCCACCAGCACCAUCCGCCACU-GACCGAU ......(((.((((((....))))))(((((((((((...........)))).((.((((((((((((.(((.....))).))))))....)))))).))..))).))))-...))). ( -39.20, z-score = -1.77, R) >anoGam1.chr3L 12325912 99 - 41284009 ----GCCACGGCAGCAGCGACGGCCCGUCGUCGUCGGAGAAGCUGAAGCGGCAGGCGAUCGACAGCUUCU--ACCAUGAUGUGGCGUGC-CGCGGGGACUACCGGG------------ ----.((.(((.(((..((.((((.(((((.((((((((((((((...((((....).))).))))))))--.))..))))))))).))-))))..).)).)))))------------ ( -42.70, z-score = -0.35, R) >consensus GCCAACUCCAAUGACGCCCACAUCGUGGUCUGGGAGAAGAAGUUAUAAUCGCAGGAGCUGGUGUGCUGCUGCACCCUUCAGCAGCAUCCCCACCAGCACCAUCCACCGCU_AACCGAG .....((((.....(((.......))).....)))).................((.(((((((((((((((.......))))))))....))))))).)).................. (-26.55 = -27.13 + 0.58)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:52:48 2011