| Sequence ID | dm3.chrX |
|---|---|
| Location | 17,334,239 – 17,334,351 |
| Length | 112 |
| Max. P | 0.943879 |

| Location | 17,334,239 – 17,334,351 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 82.68 |
| Shannon entropy | 0.29090 |
| G+C content | 0.50992 |
| Mean single sequence MFE | -24.89 |
| Consensus MFE | -16.16 |
| Energy contribution | -16.80 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.943879 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 17334239 112 - 22422827 CUCCUGCGCCACCGAGCUGUGUAUUUUCCAUUUUCCCGCUCCU-CCACUUGGAUUACUUUCGCCACCA---CAGUGGCGUUCUCCCCAAAGUGGAGUACAUGCUUCCUCAAUUAUG .....(((.....((((....................))))((-((((((((........((((((..---..))))))......)).))))))))....)))............. ( -28.09, z-score = -3.06, R) >droEre2.scaffold_4690 7673794 98 - 18748788 --------------AACUAUGUACUUUCCAUUUGGCCGCUCCU-CCAGUACCACUACUUUCACCACCA---CAGUGGCGUUCUCCCCAAAGUGGAGUACAUGCAUCCUCAAUUAUG --------------...(((((((((..(.(((((........-..((((....))))..(.((((..---..)))).)......))))))..))))))))).............. ( -20.10, z-score = -1.97, R) >droYak2.chrX 15953706 99 - 21770863 --------------UGCUAUGAAUUUUCCAUUUUCCCGCUCCUCCCACUUGCAUUACCUUCGCCACCA---CAGUGGCGUUCUCCCCAAAGUGGAGUACAUGCAUCCGCAAUUAUG --------------(((.(((....(((((((((...((...........))........((((((..---..))))))........)))))))))..))))))............ ( -21.90, z-score = -2.93, R) >droSec1.super_17 1180488 114 - 1527944 -CGCCGCGCCACCGAACUGUGUGUUUUCCAUUUUCCCGUUCCU-CCACUUGGAUUACUUUCGCCACCACCGCAGUGGCGUUCUCCCCAAAGUGGAGUACAUGCUUCCUCAAUUAUG -.(((((((.........)))))..............((..((-((((((((........((((((.......))))))......)).)))))))).))..))............. ( -27.94, z-score = -2.18, R) >droSim1.chrX 3462420 113 + 17042790 -CGCCGCGCCACCGAACUGUGUAUUUUCCAUUUUCCCGCU-CU-CCACUUGGAUUACUUUCGCCACCACCACAGUGGCGUUCUCCCCAAAGUGGAGUACAUGCUUCCUCAAUUAUG -....(((.(((......)))................(..-((-((((((((........((((((.......))))))......)).))))))))..).)))............. ( -26.44, z-score = -2.39, R) >consensus ___C_GCGCCACCGAACUGUGUAUUUUCCAUUUUCCCGCUCCU_CCACUUGGAUUACUUUCGCCACCA___CAGUGGCGUUCUCCCCAAAGUGGAGUACAUGCUUCCUCAAUUAUG ..................((((((..((((((((..........((....))........((((((.......))))))........))))))))))))))............... (-16.16 = -16.80 + 0.64)


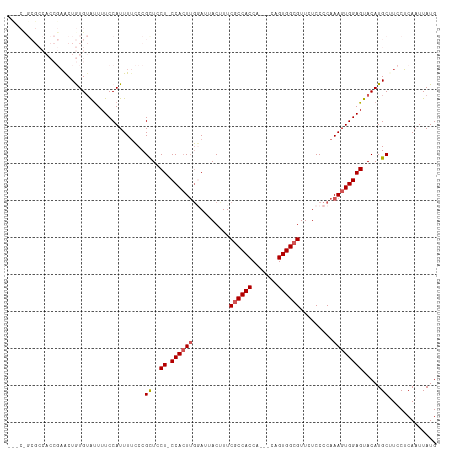
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:52:44 2011