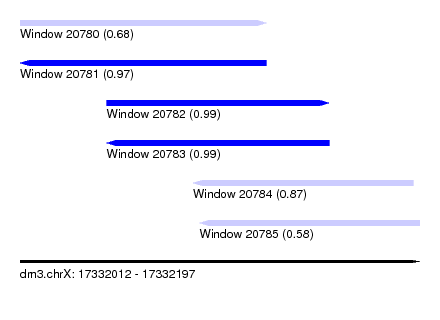
| Sequence ID | dm3.chrX |
|---|---|
| Location | 17,332,012 – 17,332,197 |
| Length | 185 |
| Max. P | 0.990912 |

| Location | 17,332,012 – 17,332,126 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 90.83 |
| Shannon entropy | 0.14872 |
| G+C content | 0.38945 |
| Mean single sequence MFE | -25.05 |
| Consensus MFE | -21.26 |
| Energy contribution | -22.20 |
| Covariance contribution | 0.94 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.85 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.679687 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
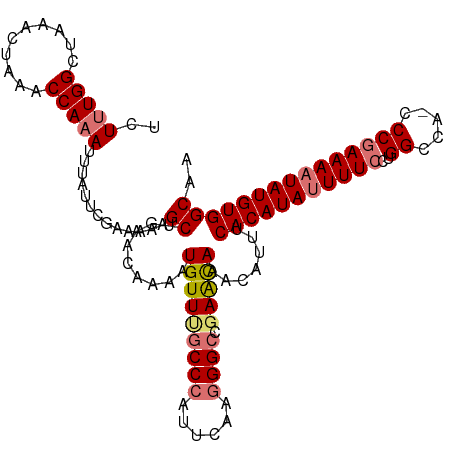
>dm3.chrX 17332012 114 + 22422827 UCUUUGGCUAAACUAAACCAAAUUUAUUCGAAAAAGCUGAACAAAAUGUUAGCCCAUUCAAGGGACGAACAAACAUUUCACAUAUUUUCCUGGCCA-CCCGAAAAUAUGUGGCAA ..(((((..........))))).............((.........((((..(((......)))...)))).......(((((((((((..((...-.))))))))))))))).. ( -23.60, z-score = -1.87, R) >droYak2.chrX 15951537 115 + 21770863 UCUUUGGCUAAACGAAACCAAAUUUAUGCGUAAAGGCUGAACAAAUUGUUCGCCCAUUCGCAGGCAGAAUAAACAUUCCACAUACUUUCCAGGCCAUCCCAAAAAUAUGUGGCAA ..(((((..........)))))....((((....(((.((((.....)))))))....)))).((.((((....))))((((((.(((...((....))..))).)))))))).. ( -23.90, z-score = -0.78, R) >droSec1.super_17 1178283 114 + 1527944 UCUUUGGCUAAACUAAACCAAAUUUAUUCGAAAAAGCUGAACAAAAUGUUUGCCCAUUCAAGGGCCGAGCAAACAUUUCACAUAUUUUCCUGGCCA-CCCGAAAAUAUGUGGCAA ..(((((..........))))).............((.........(((((((((......))).)))))).......(((((((((((..((...-.))))))))))))))).. ( -26.70, z-score = -1.98, R) >droSim1.chrX 3460174 114 - 17042790 UCUCUGGCUAAACUAAACCAAAUUUAUUCGAAAAAGCUGAACAAAAUGUUUGCCCAUUCAAGGGCCGAGCAAACAUUUCACAUAUUUUCCUGGCCA-CCCGAAAAUAUGUGGCUA ....((((...........................(((((((.....))))((((......))))..)))........(((((((((((..((...-.))))))))))))))))) ( -26.00, z-score = -1.97, R) >consensus UCUUUGGCUAAACUAAACCAAAUUUAUUCGAAAAAGCUGAACAAAAUGUUUGCCCAUUCAAGGGCCGAACAAACAUUUCACAUAUUUUCCUGGCCA_CCCGAAAAUAUGUGGCAA ..(((((..........))))).............((.........(((((((((......)))).))))).......(((((((((((..((.....))))))))))))))).. (-21.26 = -22.20 + 0.94)

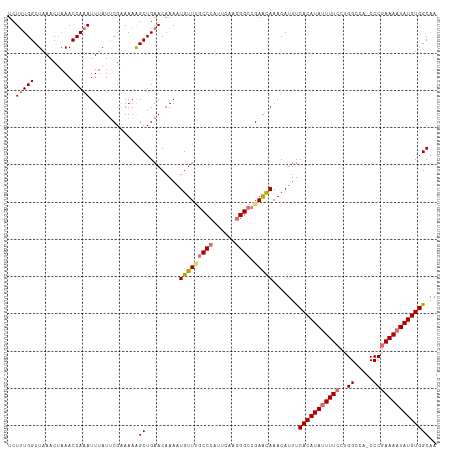
| Location | 17,332,012 – 17,332,126 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 90.83 |
| Shannon entropy | 0.14872 |
| G+C content | 0.38945 |
| Mean single sequence MFE | -34.83 |
| Consensus MFE | -29.62 |
| Energy contribution | -29.50 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.85 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.78 |
| SVM RNA-class probability | 0.967087 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
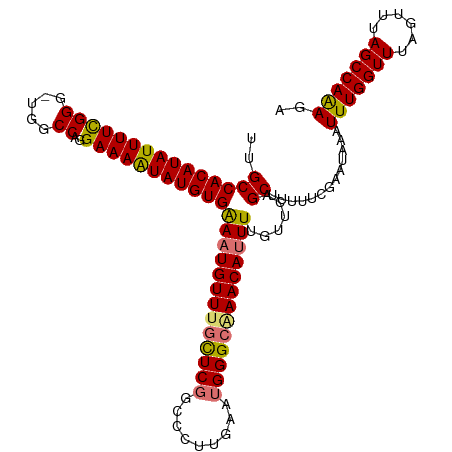
>dm3.chrX 17332012 114 - 22422827 UUGCCACAUAUUUUCGGG-UGGCCAGGAAAAUAUGUGAAAUGUUUGUUCGUCCCUUGAAUGGGCUAACAUUUUGUUCAGCUUUUUCGAAUAAAUUUGGUUUAGUUUAGCCAAAGA ....(((((((((((.((-...))..))))))))))).....((((((((..((......))((((((.....))).))).....))))))))(((((((......))))))).. ( -30.80, z-score = -1.97, R) >droYak2.chrX 15951537 115 - 21770863 UUGCCACAUAUUUUUGGGAUGGCCUGGAAAGUAUGUGGAAUGUUUAUUCUGCCUGCGAAUGGGCGAACAAUUUGUUCAGCCUUUACGCAUAAAUUUGGUUUCGUUUAGCCAAAGA ...((((((((((((.((.....)).))))))))))))...............((((...((((((((.....)))).))))...))))....(((((((......))))))).. ( -38.40, z-score = -3.06, R) >droSec1.super_17 1178283 114 - 1527944 UUGCCACAUAUUUUCGGG-UGGCCAGGAAAAUAUGUGAAAUGUUUGCUCGGCCCUUGAAUGGGCAAACAUUUUGUUCAGCUUUUUCGAAUAAAUUUGGUUUAGUUUAGCCAAAGA ....(((((((((((.((-...))..)))))))))))(((((((((((((.........)))))))))))))(((((.........)))))..(((((((......))))))).. ( -34.80, z-score = -2.40, R) >droSim1.chrX 3460174 114 + 17042790 UAGCCACAUAUUUUCGGG-UGGCCAGGAAAAUAUGUGAAAUGUUUGCUCGGCCCUUGAAUGGGCAAACAUUUUGUUCAGCUUUUUCGAAUAAAUUUGGUUUAGUUUAGCCAGAGA .((((((((((((((.((-...))..)))))))))))(((((((((((((.........)))))))))))))......)))............(((((((......))))))).. ( -35.30, z-score = -2.55, R) >consensus UUGCCACAUAUUUUCGGG_UGGCCAGGAAAAUAUGUGAAAUGUUUGCUCGGCCCUUGAAUGGGCAAACAUUUUGUUCAGCUUUUUCGAAUAAAUUUGGUUUAGUUUAGCCAAAGA ..(((((((((((((.((....))..)))))))))))(((((((((((((.........)))))))))))))......)).............(((((((......))))))).. (-29.62 = -29.50 + -0.12)

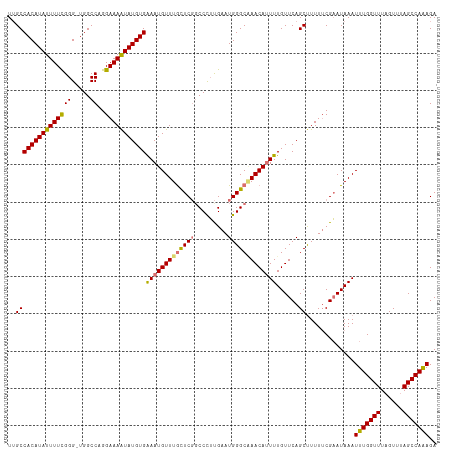
| Location | 17,332,052 – 17,332,155 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 86.22 |
| Shannon entropy | 0.21927 |
| G+C content | 0.43377 |
| Mean single sequence MFE | -30.88 |
| Consensus MFE | -27.08 |
| Energy contribution | -28.08 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.88 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.42 |
| SVM RNA-class probability | 0.990395 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 17332052 103 + 22422827 ACAAAAUGUUAGCCCAUUCAAGGGACGAACAAACAUUUCACAUAUUUUCCUGGCCA-CCCGAAAAUAUGUGGCAACAGAAACAAGU-------UGUUUGGCCAGGACACAC---- ......((((..(((......)))...))))................(((((((((-..(((.....(.((....)).)......)-------))..))))))))).....---- ( -30.40, z-score = -3.40, R) >droYak2.chrX 15951577 115 + 21770863 ACAAAUUGUUCGCCCAUUCGCAGGCAGAAUAAACAUUCCACAUACUUUCCAGGCCAUCCCAAAAAUAUGUGGCAACUGAAACAAGUAUGUCCUUGUUUGGCCAGGACACAUACAU ......(((((((......)).(((....((((((....((((((((..((((((((...........)))))..)))....))))))))...))))))))).)))))....... ( -26.90, z-score = -1.51, R) >droSec1.super_17 1178323 110 + 1527944 ACAAAAUGUUUGCCCAUUCAAGGGCCGAGCAAACAUUUCACAUAUUUUCCUGGCCA-CCCGAAAAUAUGUGGCAAUAGAAACUAGUAUGUCCUUGUUUGGCCAGGACACAC---- ......(((((((((......))).))))))................(((((((((-..(((..((((((((.........))).)))))..)))..))))))))).....---- ( -32.80, z-score = -2.69, R) >droSim1.chrX 3460214 110 - 17042790 ACAAAAUGUUUGCCCAUUCAAGGGCCGAGCAAACAUUUCACAUAUUUUCCUGGCCA-CCCGAAAAUAUGUGGCUACAGAAACAAGUAUGUCCUUGUUUGGCCAGGACACAC---- ......(((((((((......))).))))))................((((((((.-.(((........)))......(((((((......))))))))))))))).....---- ( -33.40, z-score = -2.79, R) >consensus ACAAAAUGUUUGCCCAUUCAAGGGCCGAACAAACAUUUCACAUAUUUUCCUGGCCA_CCCGAAAAUAUGUGGCAACAGAAACAAGUAUGUCCUUGUUUGGCCAGGACACAC____ ......(((((((((......)))).)))))................((((((((...(((........)))......(((((((......)))))))))))))))......... (-27.08 = -28.08 + 1.00)

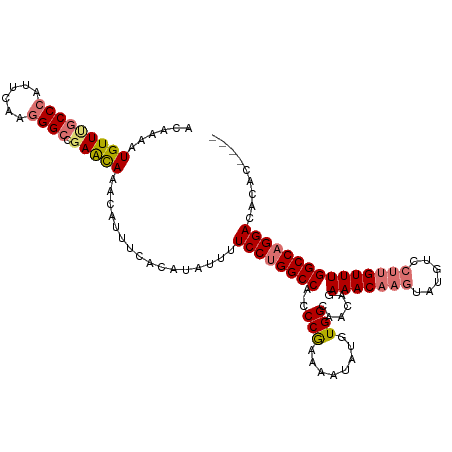
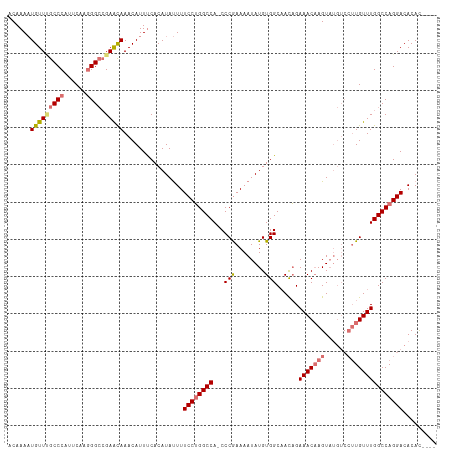
| Location | 17,332,052 – 17,332,155 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 86.22 |
| Shannon entropy | 0.21927 |
| G+C content | 0.43377 |
| Mean single sequence MFE | -37.53 |
| Consensus MFE | -30.84 |
| Energy contribution | -31.90 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.86 |
| Structure conservation index | 0.82 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.45 |
| SVM RNA-class probability | 0.990912 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 17332052 103 - 22422827 ----GUGUGUCCUGGCCAAACA-------ACUUGUUUCUGUUGCCACAUAUUUUCGGG-UGGCCAGGAAAAUAUGUGAAAUGUUUGUUCGUCCCUUGAAUGGGCUAACAUUUUGU ----((((.(((((((((..((-------((........))))((..........)).-)))))))))..))))(..(((((((.((((((.......)))))).)))))))..) ( -31.80, z-score = -2.34, R) >droYak2.chrX 15951577 115 - 21770863 AUGUAUGUGUCCUGGCCAAACAAGGACAUACUUGUUUCAGUUGCCACAUAUUUUUGGGAUGGCCUGGAAAGUAUGUGGAAUGUUUAUUCUGCCUGCGAAUGGGCGAACAAUUUGU .....(((((((((......).)))))))).((((((..(((.((((((((((((.((.....)).)))))))))))))))((((((((.......))))))))))))))..... ( -36.10, z-score = -1.88, R) >droSec1.super_17 1178323 110 - 1527944 ----GUGUGUCCUGGCCAAACAAGGACAUACUAGUUUCUAUUGCCACAUAUUUUCGGG-UGGCCAGGAAAAUAUGUGAAAUGUUUGCUCGGCCCUUGAAUGGGCAAACAUUUUGU ----((((((((((......).))))))))).............(((((((((((.((-...))..)))))))))))(((((((((((((.........)))))))))))))... ( -40.40, z-score = -3.53, R) >droSim1.chrX 3460214 110 + 17042790 ----GUGUGUCCUGGCCAAACAAGGACAUACUUGUUUCUGUAGCCACAUAUUUUCGGG-UGGCCAGGAAAAUAUGUGAAAUGUUUGCUCGGCCCUUGAAUGGGCAAACAUUUUGU ----((((.((((((((..(((..((((....))))..))).(((.(........)))-)))))))))..))))(..(((((((((((((.........)))))))))))))..) ( -41.80, z-score = -3.70, R) >consensus ____GUGUGUCCUGGCCAAACAAGGACAUACUUGUUUCUGUUGCCACAUAUUUUCGGG_UGGCCAGGAAAAUAUGUGAAAUGUUUGCUCGGCCCUUGAAUGGGCAAACAUUUUGU ....((...(((((((((((((((......)))))))......((..........))...))))))))..))..(..(((((((((((((.........)))))))))))))..) (-30.84 = -31.90 + 1.06)

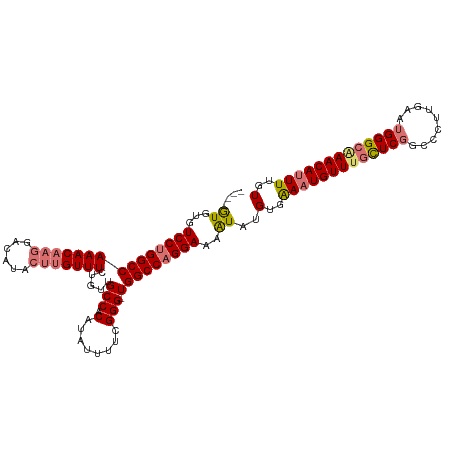
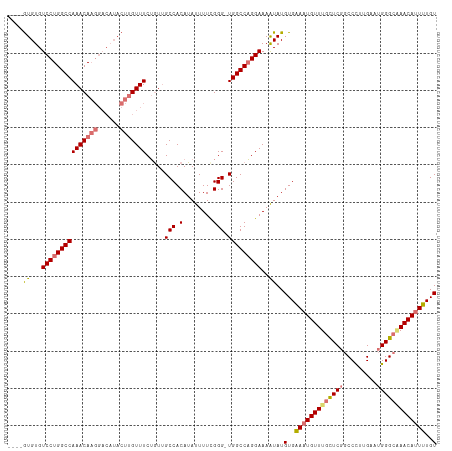
| Location | 17,332,092 – 17,332,194 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 84.82 |
| Shannon entropy | 0.23772 |
| G+C content | 0.51564 |
| Mean single sequence MFE | -35.95 |
| Consensus MFE | -25.05 |
| Energy contribution | -25.93 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.872403 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 17332092 102 - 22422827 GCCAU-GCCCUCCGCCCACCAACCGGAUUAUUCCGAGA----GUGUGUGUCCUGGCCAAACA-------ACUUGUUUCUGUUGCCACAUAUUUUCGGG-UGGCCAGGAAAAUAUG .....-..(((((((((.......(((....)))((((----(((((((.(..(((.((((.-------....))))..)))).))))))))))))))-)))..)))........ ( -31.50, z-score = -1.91, R) >droYak2.chrX 15951617 115 - 21770863 GCCACAACCGCCCUGCUAACAACCGCAUAAUUCCGAGAGUAUGUAUGUGUCCUGGCCAAACAAGGACAUACUUGUUUCAGUUGCCACAUAUUUUUGGGAUGGCCUGGAAAGUAUG .......(((((.(((........)))..(((((((((((((((.(((((((((......).))))))))...((.......)).))))))))))))))))))..))........ ( -31.70, z-score = -1.12, R) >droSec1.super_17 1178363 109 - 1527944 GCCAC-GCCCUCCGGCCGCCAGCCGGAUUAUUCCGAGA----GUGUGUGUCCUGGCCAAACAAGGACAUACUAGUUUCUAUUGCCACAUAUUUUCGGG-UGGCCAGGAAAAUAUG (((((-....((((((.....)))))).....((((((----((((((((((((......).)))))......((.......)).)))))))))))))-))))............ ( -40.70, z-score = -3.12, R) >droSim1.chrX 3460254 109 + 17042790 GCCAC-GCCCUCCGCCCGCCAGCCGGAUUAUUCCGAGA----GUGUGUGUCCUGGCCAAACAAGGACAUACUUGUUUCUGUAGCCACAUAUUUUCGGG-UGGCCAGGAAAAUAUG ..(((-((.(((.((......))((((....)))).))----).)))))((((((((..(((..((((....))))..))).(((.(........)))-)))))))))....... ( -39.90, z-score = -3.03, R) >consensus GCCAC_GCCCUCCGCCCACCAACCGGAUUAUUCCGAGA____GUGUGUGUCCUGGCCAAACAAGGACAUACUUGUUUCUGUUGCCACAUAUUUUCGGG_UGGCCAGGAAAAUAUG .......................((((....)))).......((((...(((((((((((((((......)))))))......((..........))...))))))))..)))). (-25.05 = -25.93 + 0.88)

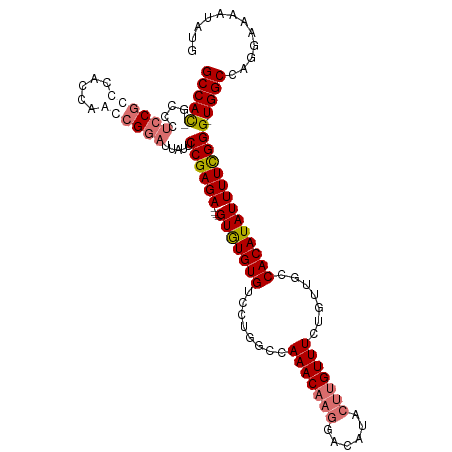
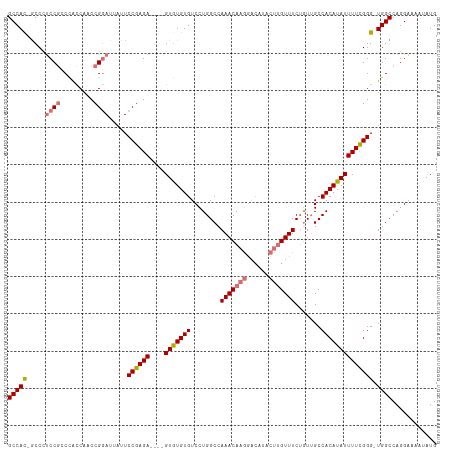
| Location | 17,332,095 – 17,332,197 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 84.38 |
| Shannon entropy | 0.24478 |
| G+C content | 0.52268 |
| Mean single sequence MFE | -36.27 |
| Consensus MFE | -22.90 |
| Energy contribution | -24.53 |
| Covariance contribution | 1.63 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.575128 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 17332095 102 - 22422827 CGAGCCAU-GCCCUCCGCCCACCAACCGGAUUAUUCCGAGA----GUGUGUGUCCUGGCCAAACA-------ACUUGUUUCUGUUGCCACAUAUUUUCGGG-UGGCCAGGAAAAU ........-..(((((((((.......(((....)))((((----(((((((.(..(((.((((.-------....))))..)))).))))))))))))))-)))..)))..... ( -31.50, z-score = -1.50, R) >droYak2.chrX 15951620 115 - 21770863 AGAGCCACAACCGCCCUGCUAACAACCGCAUAAUUCCGAGAGUAUGUAUGUGUCCUGGCCAAACAAGGACAUACUUGUUUCAGUUGCCACAUAUUUUUGGGAUGGCCUGGAAAGU ((.(((..........(((........)))..(((((((((((((((.(((((((((......).))))))))...((.......)).))))))))))))))))))))....... ( -32.30, z-score = -1.15, R) >droSec1.super_17 1178366 109 - 1527944 CGAGCCAC-GCCCUCCGGCCGCCAGCCGGAUUAUUCCGAGA----GUGUGUGUCCUGGCCAAACAAGGACAUACUAGUUUCUAUUGCCACAUAUUUUCGGG-UGGCCAGGAAAAU .(.(((((-....((((((.....)))))).....((((((----((((((((((((......).)))))......((.......)).)))))))))))))-)))))........ ( -41.40, z-score = -3.03, R) >droSim1.chrX 3460257 109 + 17042790 CGAGCCAC-GCCCUCCGCCCGCCAGCCGGAUUAUUCCGAGA----GUGUGUGUCCUGGCCAAACAAGGACAUACUUGUUUCUGUAGCCACAUAUUUUCGGG-UGGCCAGGAAAAU .....(((-((.(((.((......))((((....)))).))----).)))))((((((((..(((..((((....))))..))).(((.(........)))-))))))))).... ( -39.90, z-score = -2.71, R) >consensus CGAGCCAC_GCCCUCCGCCCACCAACCGGAUUAUUCCGAGA____GUGUGUGUCCUGGCCAAACAAGGACAUACUUGUUUCUGUUGCCACAUAUUUUCGGG_UGGCCAGGAAAAU ...............(((.(((...(((((....)))).).....))).)))(((((((((((((((......)))))))......((..........))...)))))))).... (-22.90 = -24.53 + 1.63)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:52:41 2011