
| Sequence ID | dm3.chrX |
|---|---|
| Location | 17,326,406 – 17,326,497 |
| Length | 91 |
| Max. P | 0.732264 |

| Location | 17,326,406 – 17,326,497 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 71.94 |
| Shannon entropy | 0.50857 |
| G+C content | 0.38129 |
| Mean single sequence MFE | -19.87 |
| Consensus MFE | -9.44 |
| Energy contribution | -12.30 |
| Covariance contribution | 2.86 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.732264 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
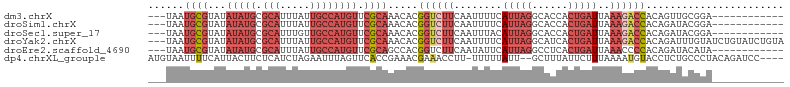
>dm3.chrX 17326406 91 + 22422827 ---UAAUGCGUAUAUAUGCGCAUUUAUUGCCAUGUUCGCAAACACGGUCUUCAAUUUUCAUUAGGCACCACUGAUUAAAGACCACAGUUGCGGA------------ ---.((((((((....))))))))..........(((((((....((((((........(((((......)))))..))))))....)))))))------------ ( -23.80, z-score = -2.13, R) >droSim1.chrX 3454450 91 - 17042790 ---UAAUGCGUAUAUAUGCGCAUUUAUUGCCAUGUUCGCAAACACGGUCUUCAAUUUUCAUUAGGCACCACUGAUUAAAGACCACAGAUACGGA------------ ---...((((...(((((.(((.....)))))))).)))).....((((((........(((((......)))))..))))))...........------------ ( -21.10, z-score = -1.92, R) >droSec1.super_17 1172668 91 + 1527944 ---UAAUGCGUAUAUAUGCGCAUUUGUUGCCAUGUUCGCAAACACGGUCUUCAAUUUACAUUAGGCACCACUGAUUAAAGACCACAGAUACGGA------------ ---...((((...(((((.(((.....)))))))).)))).....((((((.....((.(((((......)))))))))))))...........------------ ( -21.40, z-score = -1.49, R) >droYak2.chrX 15945942 103 + 21770863 ---UAAUGCGUAUAUAUGCGCAUUUAUUGCCAUGUUCGCAAACACGGUCUUCAAUUUUCAUUAGGCAUCACUGAUUAAAGACCACAGAUUUGUAUCUGUAUCUGUA ---...((((...(((((.(((.....)))))))).)))).(((.((((((........(((((......)))))..))))))((((((....))))))...))). ( -27.00, z-score = -2.87, R) >droEre2.scaffold_4690 7665689 91 + 18748788 ---UAAUGCGUAUAUAUGCGCAUUUAUUGCCAUGUUCGCAGCCACGGUCUUCAAUAUUCAUUAGGCCUCACUGAUUAAACCCCACAGAUACAUA------------ ---...((((...(((((.(((.....)))))))).)))).....(((((............)))))...(((...........))).......------------ ( -17.90, z-score = -1.03, R) >dp4.chrXL_group1e 1669874 99 + 12523060 AUGUAAUUUUCAUUACUUCUCAUCUAGAAUUUAGUUCACCGAAACGAAACCUU-UUUUUAUU--GCUUUAUUCUUUAAAAUGUACCUCUGCCCUACAGAUCC---- ..(((((....))))).........(((((..(((.....((((.(....).)-))).....--)))..)))))............((((.....))))...---- ( -8.00, z-score = -0.21, R) >consensus ___UAAUGCGUAUAUAUGCGCAUUUAUUGCCAUGUUCGCAAACACGGUCUUCAAUUUUCAUUAGGCACCACUGAUUAAAGACCACAGAUACGGA____________ ......((((...(((((.(((.....)))))))).)))).....((((((........(((((......)))))..))))))....................... ( -9.44 = -12.30 + 2.86)

| Location | 17,326,406 – 17,326,497 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 71.94 |
| Shannon entropy | 0.50857 |
| G+C content | 0.38129 |
| Mean single sequence MFE | -21.85 |
| Consensus MFE | -12.53 |
| Energy contribution | -13.45 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.43 |
| Mean z-score | -0.95 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.516492 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
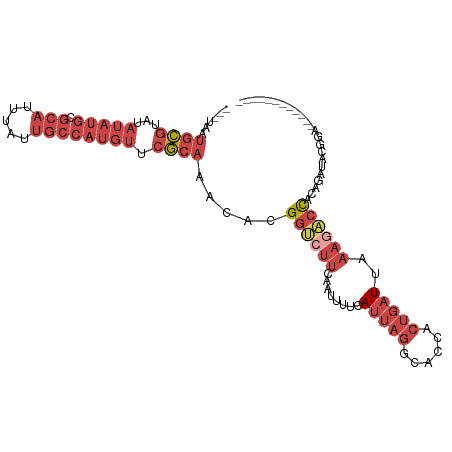
>dm3.chrX 17326406 91 - 22422827 ------------UCCGCAACUGUGGUCUUUAAUCAGUGGUGCCUAAUGAAAAUUGAAGACCGUGUUUGCGAACAUGGCAAUAAAUGCGCAUAUAUACGCAUUA--- ------------..(((((.(..((((((((((((..(....)...)))...)))))))))..).)))))............((((((........)))))).--- ( -25.60, z-score = -1.82, R) >droSim1.chrX 3454450 91 + 17042790 ------------UCCGUAUCUGUGGUCUUUAAUCAGUGGUGCCUAAUGAAAAUUGAAGACCGUGUUUGCGAACAUGGCAAUAAAUGCGCAUAUAUACGCAUUA--- ------------..((((..(..((((((((((((..(....)...)))...)))))))))..)..))))............((((((........)))))).--- ( -23.00, z-score = -1.15, R) >droSec1.super_17 1172668 91 - 1527944 ------------UCCGUAUCUGUGGUCUUUAAUCAGUGGUGCCUAAUGUAAAUUGAAGACCGUGUUUGCGAACAUGGCAACAAAUGCGCAUAUAUACGCAUUA--- ------------..((((..(..((((((((((((..(....)...))...))))))))))..)..))))....((....))((((((........)))))).--- ( -24.20, z-score = -1.25, R) >droYak2.chrX 15945942 103 - 21770863 UACAGAUACAGAUACAAAUCUGUGGUCUUUAAUCAGUGAUGCCUAAUGAAAAUUGAAGACCGUGUUUGCGAACAUGGCAAUAAAUGCGCAUAUAUACGCAUUA--- (((....((((((....))))))((((((((((...(.((.....)).)..))))))))))))).((((.......))))..((((((........)))))).--- ( -25.80, z-score = -2.14, R) >droEre2.scaffold_4690 7665689 91 - 18748788 ------------UAUGUAUCUGUGGGGUUUAAUCAGUGAGGCCUAAUGAAUAUUGAAGACCGUGGCUGCGAACAUGGCAAUAAAUGCGCAUAUAUACGCAUUA--- ------------......((..((((.((((.....)))).))))..)).(((((....(((((........))))))))))((((((........)))))).--- ( -19.50, z-score = 0.61, R) >dp4.chrXL_group1e 1669874 99 - 12523060 ----GGAUCUGUAGGGCAGAGGUACAUUUUAAAGAAUAAAGC--AAUAAAAA-AAGGUUUCGUUUCGGUGAACUAAAUUCUAGAUGAGAAGUAAUGAAAAUUACAU ----...((((.....))))......................--........-....((((((((.((..........)).)))))))).(((((....))))).. ( -13.00, z-score = 0.03, R) >consensus ____________UCCGUAUCUGUGGUCUUUAAUCAGUGAUGCCUAAUGAAAAUUGAAGACCGUGUUUGCGAACAUGGCAAUAAAUGCGCAUAUAUACGCAUUA___ ..............((((..(..((((((((((..((........))....))))))))))..)..))))............((((((........)))))).... (-12.53 = -13.45 + 0.92)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:52:36 2011